(1)
Department of Dermatology and Skin Science, University of British Columbia, Vancouver, Canada
(2)
School of Computing Science, Simon Fraser University, Vancouver, Canada
(3)
Cancer Control Research Program, BC Cancer Research Center, Vancouver, Canada
(4)
Martinos Center for Biomedical Imaging, Harvard Medical School, Boston, USA
Abstract
We describe the importance of identifying pigment networks in lesions which may be melanomas, and survey methods for identifying pigment networks (PN) in dermoscopic images. We then give details of how machine learning can be used to classify images into three classes: PN Absent, Regular PN and Irregular PN.
Keywords
Dermoscopic structuresPigment networkMelanomaComputer-aided diagnosisMachine learningGraph-based analysisIntroduction
Malignant melanoma, the most deadly form of skin cancer, is one of the most rapidly increasing cancers in the world. Melanoma is now the fifth most common malignancy in the United States [1], with an estimate of 9,180 deaths out of 76,250 incidences in the United States during 2012 [2]. Metastatic melanoma is very difficult to treat, so the best treatment is still early diagnosis and prompt surgical excision of the primary cancer so that it can be completely excised while it is still localized. Unlike many cancers, melanoma is visible on the skin; up to 70 % of all melanomas are first identified by the patients themselves (53 %) or close family members (17 %) [3]. Therefore, advances in computer-aided diagnostic methods based on digital images, as aids to self-examining approaches, may significantly reduce the mortality.
In almost all the clinical dermoscopy methods, dermatologists look for the presence of specific visual features for making a diagnosis of melanoma. Then, these features are analyzed for irregularities and malignancy [4–7]. The most important diagnostic feature of melanocytic lesions is the pigment network, which consists of pigmented network lines and hypo-pigmented holes [7]. These structures show prominent lines, homogeneous or inhomogeneous meshes. The anatomic basis of the pigment network is either melanin pigment in keratinocytes, or in melanocytes along the dermoepidermal junction. The reticulation (network) represents the rete-ridge pattern of the epidermis. The holes in the network correspond to tips of the dermal papillae and the overlying suprapapillary plates of the epidermis [8, 9].
A pigment network can be classified as either Typical or Atypical, where the definition of a Typical pigment network is: “a light-to-dark-brown network with small, uniformly spaced network holes and thin network lines distributed more or less regularly throughout the lesion and usually thinning out at the periphery” [7]. For an Atypical pigment network, the definition is: “a black, brown or gray network with irregular holes and thick lines” [7]. The goal is to automatically classify a given image to one of three classes: Absent, Typical, or Atypical.
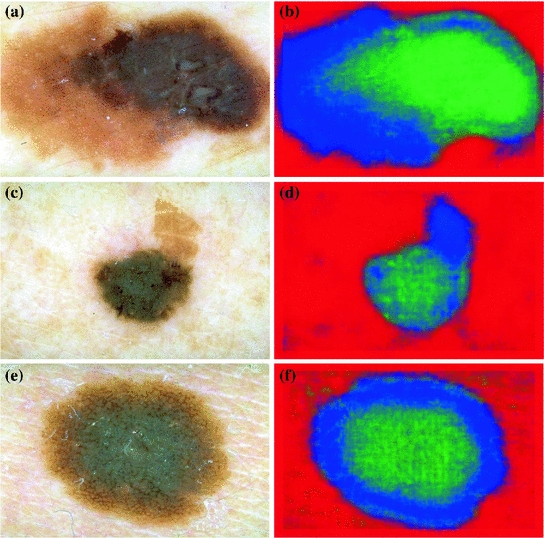
Figure 1 illustrates these three classes.
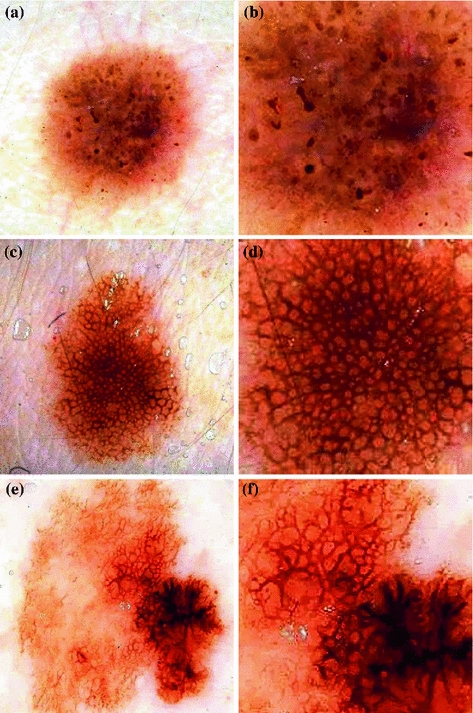

Fig. 1
The three classes of the dermoscopic structure pigment network: a–b Absent; c–d Typical; and e–f Atypical. b, d, f are magnifications of (a, c, e) respectively
In melanocytic nevi, the pigment network is slightly pigmented. Light brown network lines are thin and fade gradually at the periphery. Holes are regular and narrow. In melanoma, the pigment network usually ends abruptly at the periphery and has irregular holes, thickened and darkened network lines, and treelike branching at the periphery where pigment network features change between bordering regions [10]. Some areas of malignant lesions manifest as a broad and prominent pigment network, while others have a discrete irregular pigment network. The pigment network also may be absent in some areas or the entire lesion.
To simulate an expert’s diagnostic approach, an automated analysis of dermoscopy images requires several steps. Delineation of the region of interest, which has been widely addressed in the literature, is always the first essential step in a computerized analysis of skin lesion images [11, 12]. The border characteristics provide essential information for an accurate diagnosis [13]. For instance, asymmetry, border irregularity, and abrupt border cutoff are some of the critical features calculated based on the lesion border [14]. Furthermore, the extraction of other critical clinical indicators and dermoscopy structures such as atypical pigment networks, globules, and blue-white areas depend on the border detection. The next essential step is the detection and analysis of the key diagnostic features of specific dermoscopic structures, such as pigment networks, discussed in this chapter.
We first review published methods for computer-aided detection and analysis of pigment networks, and then provide details of a successful approach for quantifying the irregularity of these pigment networks.
Pigment Network Detection
The automated detection of pigment network has received recent attention [15–24], and there is a very recent comprehensive review of computerized analysis of pigmented skin lesions, by Korotkov and Garcia [25]. Fleming et al. [15] report techniques for extracting and visualizing pigment networks via morphological operators. They investigated the thickness and the variability of thickness of network lines; the size and variability of network holes; and the presence or absence of radial streaming and pseudopods near the network periphery. They use morphological techniques in their method and their results are purely qualitative. Fischer et al. [17] use local histogram equalization and gray level morphological operations to enhance the pigment network. Anantha et al. [18] propose two algorithms for detecting pigment networks in skin lesions: one involving statistics over neighboring gray-level dependence matrices, and one involving filtering with Laws energy masks. Various Laws masks are applied and the responses are squared. Improved results are obtained by a weighted average of two Laws masks whose weights are determined empirically. Classification of these tiles is done with approximately 80 % accuracy.
Betta et al. [19] begin by taking the difference of an image and its response to a median filter. This difference image is thresholded to create a binary mask which undergoes a morphological closing operation to remove any local discontinuities. This mask is then combined with a mask created from a high-pass filter applied in the Fourier domain to exclude any slowly modulating frequencies. Results are reported visually, but appear to achieve a sensitivity of 50 % with a specificity of 100 %.
Di Leo et. al. [20] extend this method and compute features over the ‘holes’ of the pigment network. A decision tree is learned in order to classify future images and an accuracy of 71.9 % is achieved. Shrestha et. al. [21] begin with a set of 106 images where the location of the atypical pigment network (APN) has been manually segmented. If no APN is present, then the location of the most ‘irregular texture’ is manually selected. They then compute several texture metrics over these areas (energy, entropy, etc.) and employ various classifiers to label unseen images. They report accuracies of approximately 95 %.
There are three works where supervised learning has been used to detect the dermoscopic structure pigment network [22, 24, 26]. Serrano and Acha [22] use Markov random fields in a supervised setting to classify 100 tiles (sized 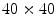 ) that have been labeled with one of five global patterns: reticular, globular, cobblestone, homogeneous, and parallel. In the context of the study, a reticular pattern can be considered equivalent to pigment network. Using tenfold cross validation, they achieve an impressive overall accuracy of 86 %, considering the difficulty of a five-class problem. It is unclear, however, how the tiles were selected. It could be the tiles were difficult, real-world examples, or that they were text-book-like definitive exemplars. Nowak et al. [24] have developed a novel method for detecting and visualizing pigment networks, based on an adaptive filter inspired by Swarm Intelligence with the advantage that there is no need to preprocess the images.
) that have been labeled with one of five global patterns: reticular, globular, cobblestone, homogeneous, and parallel. In the context of the study, a reticular pattern can be considered equivalent to pigment network. Using tenfold cross validation, they achieve an impressive overall accuracy of 86 %, considering the difficulty of a five-class problem. It is unclear, however, how the tiles were selected. It could be the tiles were difficult, real-world examples, or that they were text-book-like definitive exemplars. Nowak et al. [24] have developed a novel method for detecting and visualizing pigment networks, based on an adaptive filter inspired by Swarm Intelligence with the advantage that there is no need to preprocess the images.
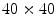 ) that have been labeled with one of five global patterns: reticular, globular, cobblestone, homogeneous, and parallel. In the context of the study, a reticular pattern can be considered equivalent to pigment network. Using tenfold cross validation, they achieve an impressive overall accuracy of 86 %, considering the difficulty of a five-class problem. It is unclear, however, how the tiles were selected. It could be the tiles were difficult, real-world examples, or that they were text-book-like definitive exemplars. Nowak et al. [24] have developed a novel method for detecting and visualizing pigment networks, based on an adaptive filter inspired by Swarm Intelligence with the advantage that there is no need to preprocess the images.
) that have been labeled with one of five global patterns: reticular, globular, cobblestone, homogeneous, and parallel. In the context of the study, a reticular pattern can be considered equivalent to pigment network. Using tenfold cross validation, they achieve an impressive overall accuracy of 86 %, considering the difficulty of a five-class problem. It is unclear, however, how the tiles were selected. It could be the tiles were difficult, real-world examples, or that they were text-book-like definitive exemplars. Nowak et al. [24] have developed a novel method for detecting and visualizing pigment networks, based on an adaptive filter inspired by Swarm Intelligence with the advantage that there is no need to preprocess the images.Wighton’s method, published in [26], used machine learning to analyze a dataset of 734 images from [8] and classify the images into Absent/Present. Labels of either Absent or Present for the structure pigment network are derived from the atlas. A custom training set is created consisting of 20 images where the pigment network is present across the entire lesion and 20 images absent of pigment network. Pixels are assigned a label from the set 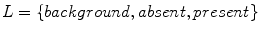 as follows: for each image, pixels outside the segmentation are assigned the label background, while pixels inside the segmentation are assigned either the label absent or present. By considering these three labels, they simultaneously segment the lesion and detect the structure pigment network. A feature-set consisting of Gaussian and Laplacian of Gaussian filter-banks was employed. They present visual results in Fig. 2 by plotting
as follows: for each image, pixels outside the segmentation are assigned the label background, while pixels inside the segmentation are assigned either the label absent or present. By considering these three labels, they simultaneously segment the lesion and detect the structure pigment network. A feature-set consisting of Gaussian and Laplacian of Gaussian filter-banks was employed. They present visual results in Fig. 2 by plotting 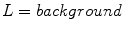 ,
, 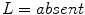 , and
, and 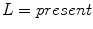 in the red, green, and blue channels, respectively.
in the red, green, and blue channels, respectively.
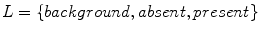 as follows: for each image, pixels outside the segmentation are assigned the label background, while pixels inside the segmentation are assigned either the label absent or present. By considering these three labels, they simultaneously segment the lesion and detect the structure pigment network. A feature-set consisting of Gaussian and Laplacian of Gaussian filter-banks was employed. They present visual results in Fig. 2 by plotting
as follows: for each image, pixels outside the segmentation are assigned the label background, while pixels inside the segmentation are assigned either the label absent or present. By considering these three labels, they simultaneously segment the lesion and detect the structure pigment network. A feature-set consisting of Gaussian and Laplacian of Gaussian filter-banks was employed. They present visual results in Fig. 2 by plotting 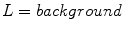 ,
, 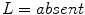 , and
, and 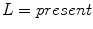 in the red, green, and blue channels, respectively.
in the red, green, and blue channels, respectively.
Fig. 2
Qualitative results of pigment network detection from [26]. First column original dermoscopic images. Second column red, green, and blue channels encode the likelihood that a pixel is labeled as background, absent, and present, respectively
Although these studies have made significant contributions, there has yet to be a comprehensive analysis of pigment network detection on a large number of dermoscopic images. All other work to date has either: (1) not reported quantitative validation [15, 17, 24, 26]; (2) validated against a small ( ) number of images [19]; (3) only considered or reported results for the 2-class problem (e.g. Absent/Present rather than Absent/Typical/Atypical) [18–21, 26]; (4) not explicitly identified the location of the network [18]; or (5) has made use of unrealistic exclusion criteria and other manual interventions [21].
) number of images [19]; (3) only considered or reported results for the 2-class problem (e.g. Absent/Present rather than Absent/Typical/Atypical) [18–21, 26]; (4) not explicitly identified the location of the network [18]; or (5) has made use of unrealistic exclusion criteria and other manual interventions [21].
 ) number of images [19]; (3) only considered or reported results for the 2-class problem (e.g. Absent/Present rather than Absent/Typical/Atypical) [18–21, 26]; (4) not explicitly identified the location of the network [18]; or (5) has made use of unrealistic exclusion criteria and other manual interventions [21].
) number of images [19]; (3) only considered or reported results for the 2-class problem (e.g. Absent/Present rather than Absent/Typical/Atypical) [18–21, 26]; (4) not explicitly identified the location of the network [18]; or (5) has made use of unrealistic exclusion criteria and other manual interventions [21].Now we describe our successful approach to analyze the texture in dermoscopy images, to detect regular and irregular pigment networks in the presence of other structures such as dots and globules. The method is based on earlier work on the 2-class problem (Absent and Present) published in [27, 28] and our work on the 3-class problem (Absent, Typical, and Atypical) published in [29] (see Fig. 1).
Pigment Network Analysis: Overview
We subdivide the structure into the darker mesh of the pigment network (which we refer to as the ‘net’) and the lighter colored areas the net surrounds (which we refer to as the ‘holes’). After identifying these substructures we use the clinical definitions previously mentioned to derive several structural, geometric, chromatic and textural features suitable for classification. The result is a robust, reliable, automated method for identifying and classifying the structure pigment network. Figure 3 illustrates an overview of our approach to irregular pigment network detection. After pre-processing, we find the ‘hole mask’ indicating the pixels belonging to the holes of the pigment network. Next, a ‘net mask’ is created, indicating the pixels belonging to the net of the pigment network. We then use these masks to compute a variety of features including structural (which characterizes shape), geometric (which characterizes distribution and uniformity), chromatic and textural features. These features are fed into a classifier to classify unseen images into three classes of Absent, Typical and Atypical. The major modules in Fig. 3 are explained in the following sub-sections.
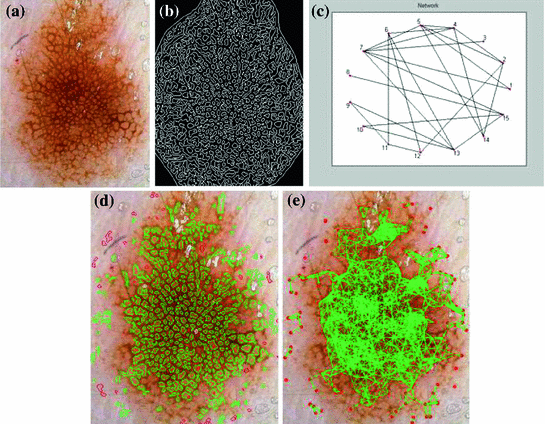
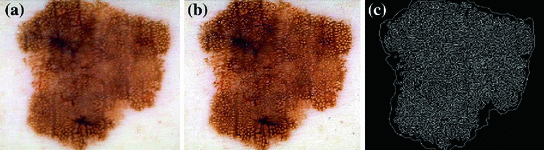

Fig. 3
Steps of the proposed algorithm for hole detection. a Original image. b LoG response. c Image to graph conversion. d Cyclic subgraphs. e Graph of holes

Fig. 4
a A given skin lesion image. b Sharpened image. c Result of the edge detection after segmenting the lesion
Pre-processing
In order to prevent unnecessary analysis of the pixels belonging to the skin, the lesion is first segmented. Either manual segmentation or our automatic segmentation method [12] was used. Next the image is sharpened using the MATLAB Image Processing Tool Box function Unsharp mask, one of the most popular tools for image sharpening [30]. A two-dimensional high-pass filter is created using Eq. 1. This high-pass filter sharpens the image by removing the low frequency noise. We use the default parameters of MATLAB in our experiments (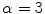 ). Figure 4b shows the result of the sharpening step.
). Figure 4b shows the result of the sharpening step.
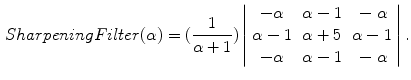
To investigate structures of the skin texture, it was necessary to reduce the color images to a single plane before applying our algorithm. Various color transforms (NTSC, L*a*b, Red, Green, and Blue channels separately, Gray(intensity image), etc) were investigated for this purpose. After the training and validation step, we selected the green channel as the luminance image. Results of the different color transformations are reported in the result section of this chapter.
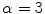 ). Figure 4b shows the result of the sharpening step.
). Figure 4b shows the result of the sharpening step.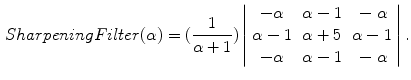
(1)
Hole Detection
As discussed previously, a pigment network is composed of holes and nets. We first describe the detection of the holes. Figure 3 shows steps of our novel graph-based approach to hole detection. After the pre-processing step described above, sharp changes of intensity are detected using the Laplacian of Gaussian (LoG) filter. The result of this edge detection step is a binary image which is subsequently converted into a graph to find holes or cyclic structures of the lesion. After finding loops or cyclic subgraphs of the graph, noise or undesired cycles are removed and a graph of the pigment network is created using the extracted cyclic structures. According to the density of the pigment network graph, the given image can be classified into Present or Absent classes, but for irregularity analysis we also need to extract more features and characteristics of the net of the network.
We used the LoG filter to detect the sharp changes of intensity along the edge of the holes inside the segmented lesion. Because of the inherent properties of the filter, it can detect the “light-dark-light” changes of the intensity well. Therefore it is good choice for blob detection and results in closed contours. The detection criterion of the edge of a hole is set to the zero crossing in the second derivative with the corresponding large peak in the first derivative. We follow the MATLAB implementation of the LoG edge detection which looks for zero crossings and their transposes. All zeros are kept and edges lie on the zero points. If there is no zero, an edge point is arbitrarily chosen as a negative second derivative point. Therefore when all ”zero” responses of the filtered image are selected, the output image includes all closed contours of the zero crossing locations inside a segmented lesion. An example of the edge detection step is shown in Figs. 4c and 3b. This black and white image captures the potential holes of the pigment network.
Now, we consider the steps necessary to extract the holes accurately. In previous works [15, 16, 19], these structures usually are found by morphologic techniques and a sequence of closing and opening functions applied to the black and white image. We did not use this approach because using morphologic techniques is error-prone in detecting the round shaped structures. Instead, the binary image is converted to a graph ( ) using 8-connected neighbors. Each pixel in the connected component is a node of
) using 8-connected neighbors. Each pixel in the connected component is a node of  and each node has a unique label according to its coordinate.
and each node has a unique label according to its coordinate.
 ) using 8-connected neighbors. Each pixel in the connected component is a node of
) using 8-connected neighbors. Each pixel in the connected component is a node of  and each node has a unique label according to its coordinate.
and each node has a unique label according to its coordinate.To find round texture features (i.e. holes), all cyclic subgraphs of 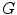 are detected using the Iterative Loop Counting Algorithm (ILCA) [31]. This algorithm transforms the network into a tree and does a depth first search on the tree for loops.
are detected using the Iterative Loop Counting Algorithm (ILCA) [31]. This algorithm transforms the network into a tree and does a depth first search on the tree for loops.
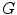 are detected using the Iterative Loop Counting Algorithm (ILCA) [31]. This algorithm transforms the network into a tree and does a depth first search on the tree for loops.
are detected using the Iterative Loop Counting Algorithm (ILCA) [31]. This algorithm transforms the network into a tree and does a depth first search on the tree for loops.After finding cyclic subgraphs which may represent the holes of a pigment network, these subgraphs were filtered and noise or wrongly detected structures (globules and dots) were removed according to parameters learned in a training and validation step.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


