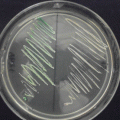
Fig. 11.1
Map of rDNA locus from Trichophyton rubrum
The region ITS1 can be amplified from a wide variety of fungus by using the primers ITS1 (5-TCCGTAGGTGAACCTTGCGG-3) and ITS2 (5-GCTGCGTTCTTCATCGATGC-3). A second variable zoned among the cluster of ribosomal DNA has an area named domain D1/D2, located in the rDNA 28S subunit. This region can be amplified with ITS1 primers (5-TCCGTAGGT GAACCTTGCGG-3) and ITS4 (5-TCCTCCGCTTATTGATATGC-3) (Fig. 11.2). They are highly conserved and informative and much less variable than the ITS. The conserved region of the domains D1/D2 is the anchorage point for the primers that, along with the variable nature of the ITS regions, provides a specific combination for each species. One of the main advantages of the ribosomal locus in fungi is the high number of copies from the target gene, 10–100 times, compared to genes with only one copy, what in the end translates in higher sensitivity [11, 19, 22, 30–33].
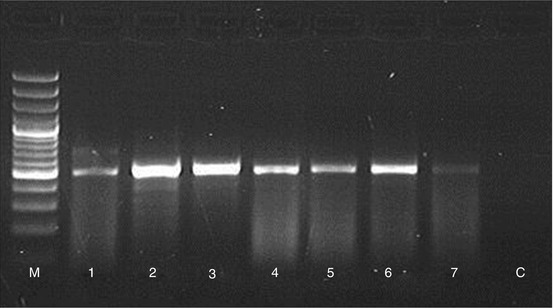

Fig. 11.2
The 18S-ITS1-5.8S-ITS2-28S rDNA amplification using the ITS1 and ITS4 primers of Trichophyton rubrum from nail samples. M, 100 bp DNA ladder; line 1, T. rubrum positive control; lines 2–7, DNA from nails with onychomycosis; C, negative control
A gene frequently used is the chitin synthase 1 (CHS1), and in cases where the ribosomal target is not sensitive enough for discriminating between closely related species, additional loci may be sequenced, such as the β-tubulin [11, 19, 22, 30–33].
More recently, real-time PCR (quantitative PCR [qPCR]) has been used to identify onychomycosis, using molecular beacons which are small single-chain probes of hairpin type that fluoresce when linked to the target site. The same way as with conventional PCR (end point PCR), both variable and conserved regions (ITS1-2/ITS1-4) can be used to design the probe. This will allow the universal identification of the fungal presence in the sample or the involved species [34].
On the other hand, in the same qPCR platform, TaqMan technology or hybrid probes can be used, such as the carboxyfluorescein (FAM)-labeled probe with a Black Hole Quencher (BHQ1) [35, 36]. This technique can detect and quantify quickly the nucleic acids directly from human and animal tissue samples. It has high sensitivity with a detection threshold of one single molecule and depends from one efficient DNA extraction and purification to avoid PCR inhibitors as well as an adequate fungal cell wall lysis [11, 12].
Panfungal Amplification Technique
The gene regions in the SSU and the LSU rDNA have highly conserved regions and variable regions [31, 37]. These permit the development of panfungal primers based on the conserved regions of the rRNA cluster which are capable of identifying a great number of fungal species and can even be species specific [27, 29, 31]. These characteristics have made this the preferred PCR mode. After the amplification of the mentioned region, different genders and species can be identified using the same amplification product by RFLP, hybridization with specific probes marked with radioactivity or digoxigenin, and, with highly specific tool, the direct sequencing of the amplification product [38–41].
A general problem with PCR as detection method for the causative agents of onychomycosis is the lack of worldwide standardization, as well as the deficit of availability of commercial systems in some countries. Several studies report excellent results with “in-house PCR”; however, the majority of these don’t make adequate comparisons mainly in clinical practice [22, 42]. The DNA extraction methods, genetic markers, as well as the different types of clinical samples (blood, DNA from fungal culture, nails, the hair, the skin, or fluids) are factors that influence the comparison between different PCR protocols.
Outlook: Future Developments
Molecular biology is a tool under continuous development and improvement for onychomycosis diagnosis.
The challenges ahead are:
Availability of molecular tests to reduce the time consumed in some techniques such as PCR sequencing, corroborate sensitivity, and specificity of the currently available tests and improve them if possible with the aim to reduce the clinical sample required that at present must be large
Improvement of keratin extraction and fungal cell wall rupture techniques
Reduction of costs
Readily accessible kits for fast etiological identification in onychomycosis, particularly when caused by non-dermatophyte molds or yeasts, which enables the indication of adequate treatment options
Improvement of subspecies identification
More than one fungal determination in the same clinical sample
Differentiation of saprophytes from etiological agents
Performance of epidemiological studies to redefine onychomycosis frequency and its etiological agents
Implementation of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS) in clinical samples
Identification of the species of causative agents with PCR-terminal restriction fragment length polymorphism (PCR-TRFLP)
Implementation of the use of other operons as genes that codify for non-ribosomal proteins and determine if relapses are by the same causative agent
Summary for the Clinician
Molecular techniques are particularly useful for fast identification of onychomycosis in atypical or mycological negative cases, gender and species identification in 24–48 h allowing a prompt diagnosis, and thus optimal management.
Clinical Pearls
Consider molecular biology for diagnosis in difficult onychomycosis cases:
Identify fungal genus and species in onychomycosis if traditional techniques have failed to do so.
Send enough of the clinical sample for adequate processing.
Contact a reference center that has access to molecular biology for difficult onychomycosis cases.
References
1.
2.
3.
4.
Ling M, Merante F, Robinson BH. A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res. 1995;23(23):4924–5.CrossRefPubMedPubMedCentral
5.
Williamson EC, Leeming JP, Palmer HM, Steward CG, Warnock D, Marks DI, Millar MR. Diagnosis of invasive aspergillosis in bone marrow transplant recipients by polymerase chain reaction. Br J Haematol. 2000;108(1):132–9.CrossRefPubMed
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree