Epidermal Growth and Differentiation: Introduction
|
Introduction
Although thin, human skin is a marvelously resilient and multifunctional organ. It performs immunomodulatory and thermoregulatory functions, is involved in social, cultural, and reproductive behaviors, and provides broad protection against water loss and environmental insults such as trauma, infection, and exposure to radiation or chemicals. The outermost layer of skin, termed the epidermis, consists of a stratified squamous epithelium and its appendages, including hair follicles, sebaceous, apocrine, and eccrine glands. This chapter discusses epidermal differentiation, with the primary focus placed on keratin filaments that are formed as major structural elements within the epidermis. Defects in epidermal keratins are known to play key roles in a number of important blistering epidermal diseases. Additional major epidermal differentiation markers, including keratohyalin granules and the cornified envelope, are also discussed.
Keratins and Epidermal Differentiation
Keratins (also known as cytokeratins) are structural proteins that belong to the superfamily of intermediate filament (IF) proteins. They are heterogeneous in size (40–70 kDa) and charge (pI 4.7–8.4), and notoriously insoluble. Sequencing the human genome revealed the presence of 54 functional keratin genes that are nearly perfectly conserved in other mammals.1 The tremendous diversity of keratin genes had not been fully appreciated until the advent of database mining and genomics, and could not be accommodated in the original nomenclature system aptly devised by Roland Moll, Werner Franke and colleagues in 1982.2 In 2006, an international effort culminated in a revised nomenclature (Table 46-1) that accommodates the newly discovered keratins, adheres to the guidelines of the Human and Mouse Gene Organization Gene Nomenclature Committee, and maintains the original designation of keratins devised by Moll and colleagues.1
Type I Keratins | Type II Keratins | ||||
|---|---|---|---|---|---|
Old Name | New Name | Main Site(s) of Expression | Old Name | New Name | Main Site(s) of Expression |
K9 | K9 | Epidermis (suprabasal) | K1 | K1 | Epidermis (suprabasal) |
K10 | K10 | Epidermis (suprabasal) | K2e | K2 | Epidermis (suprabasal) |
K12 | K12 | Cornea | K3 | K3 | Cornea (suprabasal) |
K13 | K13 | Oral mucosa | K4 | K4 | Oral mucosa (suprabasal) |
K14 | K14 | Complex epithelia | K5 | K5 | Complex epithelia (basal layer) |
K15 | K15 | Complex epithelia | K6a | K6a | Epithelial appendages |
K16 | K16 | Epithelial appendages | K6b | K6b | Epithelial appendages |
K17 | K17 | Epithelial appendages | K6e/h | K6c | Skin (needs confirm.) |
K18 | K18 | Simple epithelia | K7 | K7 | Simple epithelia |
K19 | K19 | Broad distribution | K8 | K8 | Simple epithelia |
K20 | K20 | Gut epithelium | |||
K23 | K23 | Pancreas (needs confirm.) | |||
K24 | K24 | unknown | |||
K25irs-4 | K25–K28 | Inner root sheath (hair follicles) | K6irs1–4 | K71–K74 | Inner root sheath (hair follicles) |
K6hf | K75 | Companion layer (hair follicles) | |||
K2p | K76 | Oral mucosa | |||
K1b | K77 | Sweat gland ducts | |||
K5b | K78 | Tongue | |||
K6l | K79 | Skin | |||
Kb20 | K80 | Tongue | |||
Ha1–Ha8 | K31–K38 | Hair shaft (Hair follicle) | Hb1–Hb6 | K81–K86 | Hair shaft (hair follicles) |
K39 | K39 | Hair shaft (Hair follicle) | |||
K40 | K40 | Hair shaft (Hair follicle) | |||
Sequence homology and gene substructure (number and position of introns) reveal two distinct groups of keratins of roughly equal size, designated type I and II IF genes1,3 (Fig. 46-1A). In Homo sapiens, functional type I and type II keratin genes are clustered on the long arms of chromosomes 17 (Fig. 46-1B) and 12 (Fig. 46-1C), respectively.1,4 Keratin genes are highly conserved across mammals, at the level of their organization, structure, sequence, and regulation.5 Mature filaments contain type I and II keratins in a 1:1 molar ratio.3,6 This requirement underlies the coordinated transcription of type I and II keratin genes. Remarkably, most type I and II keratin genes are regulated in a pairwise, tissue type-related, and differentiation-related fashion.7–9 This is illustrated particularly well in stratified epithelia such as epidermis (Fig. 46-2). Given their large number, differential regulation, and ease of detection (owing to abundance), keratin mRNAs and proteins represent unparalleled markers for staging the fate and differentiation of epithelial cells, under healthy and diseased conditions.
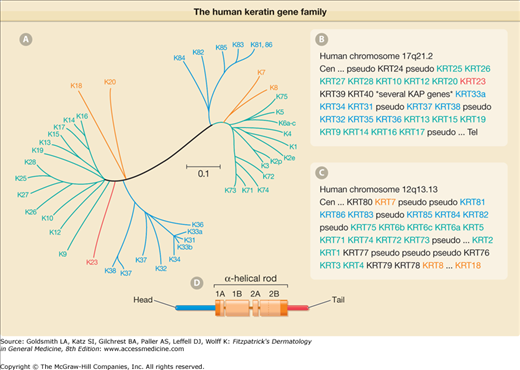
Figure 46-1
The human keratin gene family. A. Comparison of the primary structure of human keratins using the publicly available ClustalW and TreeView software. Sequence relatedness is inversely correlated with the length of the lines connecting the various sequences, and to the number and position of branch points. This comparison makes use of the sequences from the head and central rod domain for each keratin. A few keratins were left out for clarity purposes. Two major branches are seen in this tree display, corresponding to type I and type II sequences. Beyond this dichotomy, each subtype is further segregated into major subgroupings (denoted by different colors). B. Organization of functional type I keratin genes, all which are clustered on human chromosome 17, with the only exception being K18 (see “*” in C), which is located at the telomeric (Tel) boundary of the type II gene cluster. (Cen = centromere.) A large number of genes encoding keratin-associated proteins (KAP) interrupts the type I gene cluster, between KRT40 and KRT33A. [Pseudo = pseudogene (nonfunctional).] C. Organization of functional type II keratin genes, which are clustered on human chromosome 12. The K8 and K18 genes are separated by 450,000 bp. D. Schematic representation of the tripartite domain structure shared by all keratin and other IF proteins. A central α-helical “rod” domain acts as the major determinant of self-assembly. This rod domain is partitioned into subdomains 1A, 1B, 2A, and 2B, and flanked by nonhelical “head” and “tail” domains at the N– and C-termini, respectively. Both ends of the rod domain contain 15–20 amino acid regions (red) that are highly conserved among all IF proteins.
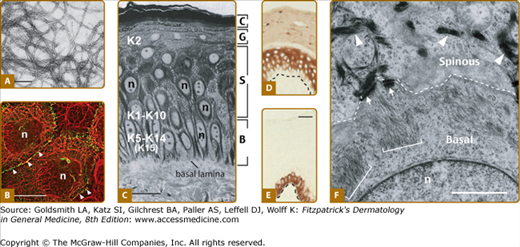
Figure 46-2
Keratin filaments and interfollicular epidermis. A. Visualization of filaments, reconstituted in vitro from purified human K5 and K14, by negative staining and electron microscopy. (Bar = 150 nm.) B. Double-labeling for keratin (red chromophore) and desmoplakin, a desmosome component (green chromophore), by indirect immunofluorescence of human epidermal cells in culture. Keratin IFs are organized in a network that spans the entire cytoplasm and are attached at desmosomal cell–cell contacts (arrowheads) between cells. (n = nucleus; bar = ∼50 μm.) [Micrograph used with permission from Dr Kathleen Green (Northwestern University).] C. Histological cross section of resin-embedded human trunk epidermis, revealing the basal (B), spinous (S), granular (G), and cornified (C) cell layers. (Bar = ∼50 μm; n = nucleus.) D and E. Differential distribution of keratin epitopes on human skin tissue cross sections as visualized by an antibody-based detection method. D. K10 is primarily concentrated in the differentiating, suprabasal layers of epidermis. E. K14 occurs in the basal layer, where the epidermal progenitor cells reside. Dashed line indicates the basal lamina. (Bar = ∼50 μm.) F. Ultrastructure of the boundary between the basal and suprabasal cells in mouse trunk epidermis, as seen by routine transmission electron microscopy. The sample, from which this micrograph was taken, is oriented in the same manner as frame C. Organization of keratin filaments as loose bundles correlates with the expression of K5–K14 in basal cells (brackets), whereas the formation of denser, electron-dense filament bundles reflects the onset of K1–K10 expression in early differentiating cells (arrowheads). Arrows point to desmosomes connecting the two cells. (Bar = 2 μm; n = nucleus.)
Despite sequence differences, all keratins display the tripartite domain structure that is typical of IF-forming proteins (Fig. 46-1D). The central domain consists of an extended α helix featuring long-range heptad repeats that mediate coiled-coil dimerization. This “rod” domain is ∼310 amino acids long and is flanked by highly variable sequences at the N– terminal head and C-terminal tail domains (Fig. 46-1D).3 Neither terminal domain exhibits known functional motifs other than the glycine loops seen in epidermal keratins.10 The head and tail domains are readily protease-accessible at the surface of the filament, where they can foster interactions with neighboring filaments, other proteins, or serve as substrates for posttranslational modifications involved in their regulation.11 Given their heterogeneity of size and primary structure, the head and tail domains are expected to make key contributions to the differential function and regulation of keratin proteins in vivo.12
The central rod domain of keratins is the main determinant of self-assembly, with important contributions from the head domain as well.13 Assembly begins with the formation of heterodimers in which the α-helical central rod domains of type I and II keratins are aligned in parallel and perfect register. Heterodimers interact along their lateral surfaces and in an end-to-end fashion to give rise to the 10–12-nm wide filaments (Fig. 46-2A) which, depending on IF protein type and assembly conditions, may contain a variable number of subunits in cross section.13 Mature IFs lack a structural polarity, a direct consequence of the antiparallel orientation of their constituent coiled–coiled dimers. The extraordinary stability of keratin subunits reflects the tightness of interactions between type I and type II keratins.14 Most of the intracellular pool of keratin proteins (>95%) is polymerized.4 There is evidence that keratin IF assembly is initiated at the cell periphery, near the cortical F-actin cytoskeleton, in cultured epithelial cells.15
Keratins form the major IF network in all epithelial cells.2,3,9 The abundance and organization of keratin IFs in vivo differ among epithelia. Keratin proteins are highly abundant (10%–80% of total cellular proteins) in surface-exposed stratified squamous epithelia (e.g., epidermis, oral mucosa, corneal epithelium, etc.).7 In epithelial cells of such tissues, keratin IFs are organized in a pancytoplasmic network extending from the surface of the nucleus to the cytoplasmic periphery, where they are membrane-anchored at sites of cell–matrix and cell–cell adhesion (hemidesmosomes, desmosomes) (Fig. 46-2B). In simple epithelia (e.g., liver, gut, pancreas, etc.), keratins are less abundant. In such tissues, polarized epithelial cells often feature asymmetrically organized keratin IFs concentrated mostly at the cytoplasmic periphery and, particularly, the apical pole.4 Several associated proteins contribute to the organization and regulation of keratin IFs in these various settings.16 Some of these proteins promote the bundling of keratin IFs (e.g., filaggrin, trichohyalin), their association with microtubules and actin microfilaments (e.g., plectin, BPAG isoforms) and/or with desmosomes or hemidesmosomes (desmoplakin, plakophilin, BPAG isoforms, etc.) (Fig. 46-2). Other partners, for example, TRADD, 14–3-3, Akt, reflect the newly discovered participation of keratin IFs in signaling roles.17
Skin provides a beautiful example of the tight relationship that has evolved between keratin gene regulation and epithelial differentiation. More than half of all known keratin genes are expressed in mature mammalian skin tissue alone. The architectural complexity of adult skin epithelia is achieved through a temporally and spatially regulated expression of keratin genes and a number of other epithelial differentiation-related genes.7,18 In the clinical setting, keratin typing is often exploited in diagnosing cancer type, its differentiation status (and therefore prognosis), as well as the origin of the cells forming metastatic foci. This strategy is also applied for diseases other than cancer (see below).4
In “thin” interfollicular epidermis (e.g., trunk; Fig. 46-2), mitotically active cells of the basal layer act as progenitors, and consistently express K5 and K14 as their main keratin pair, along with low levels of K15. Onset of differentiation coincides with the appearance of the K1/K10 pair through a robust transcriptional induction that occurs at the expense of the K5/K14 genes, which are downregulated.7,18 Accordingly, K1/K10 keratins are readily detectable in the lowermost suprabasal layer of epidermis (Fig. 46-2D). The appearance of K1 and K10 correlates with a sudden and dramatic shift in the organization of keratin IFs, which now exhibit significant bundling.19 Another type II gene, K2e, is expressed at a later stage of differentiation, i.e., the granular layer.20
The epidermis of palm and sole skin is specialized for resisting a high degree of mechanical stress, and thus is markedly thicker. This function is reflected in its architecture of alternating stripes of primary and secondary ridges,21 and again, in keratin expression. In the thick, stress-bearing, primary ridges, the major differentiation-specific (type I) K9 is presumed to foster a more resilient cytoskeleton. In the thinner secondary ridges, postmitotic keratinocytes preferentially express the (type II) K6a and (type I) K16 and K17.22 Relative to K1, K9, and K10, the properties of K6a, K16, and K17 likely foster greater cellular pliability, thereby providing flexible “hinge” regions between the more rigid, K1/K9-rich primary ridges.22 While this attractive model remains to be supported by direct experimentation, it is consistent with the dramatic upregulation of K6a, K6b, K16, and K17 that occurs in keratinocytes recruited from wound margins to participate in the restoration of the epidermal barrier following injury.23,24
Epidermal disease states are often accompanied by a deviation from normal terminal differentiation and, not surprisingly, they are almost always accompanied by altered keratin gene expression. For instance, K6a, K6b, K16, and/or K17, normally restricted to wound repair in trunk epidermis, are ectopically induced in psoriasis and related hyperproliferative disorders, nonmelanoma skin cancers, viral infections, and other conditions accompanied by inflammation.23,25,26 Similar “replacement” of the K1 and K10 keratin pair by K6, K16
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree