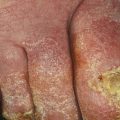
Fig. 8.1
Paronychia
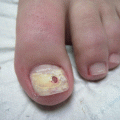
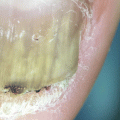
Other clinical findings comprise fungal melanonychia (Fig. 8.2), with a greenish, brownish, or even black nail discoloration [23].
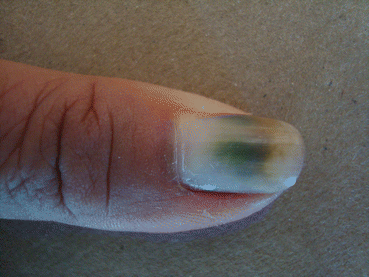

Fig. 8.2
Fungal melanonychia due to Candida
Diagnostic Clues
The diagnostic route of onychomycosis (caused by dermatophytes, Candida species, or other molds or yeasts) includes a complete interrogatory based on the patients’ history: a physical examination and microscopy and culture of nail specimens [5].
The clinical presentation of dystrophic nails (with or without discoloration of the nail plate) should alert the clinician to the possibility of onychomycosis; however, it’s necessary to use appropriate diagnostic techniques including direct microscopy and fungal culture to ensure correct diagnosis and treatment. The clinical appearance of the nail will help to recognize fungal from nonfungal etiologies of nail dystrophies. A detailed interrogation of patient’s history can help to obtain information about predisposing factors for onychomycosis such as diabetes mellitus, older age, hyperhidrosis, professional occupations, hobbies, onychogryphosis, nail trauma, poor peripheral circulation, and immunosuppression [3].
Direct examination of a nail sample with potassium/sodium hydroxide (KOH or NaOH) or Chlorazol Black is the first step to support a clinical diagnosis of onychomycosis. The specimen is obtained by scraping of nail plate in its ventral portion (hyponychium), and it should be divided into two portions for direct microscopy and culture. To identify the causative agent, the laboratory culture of sampled material is necessary to identify the specific etiologic agent.
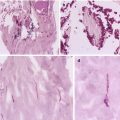
The specimen can be mounted in a solution of 20–25 % KOH or NaOH mixed with dimethyl sulfoxide and examined, first under ×10 and after under ×40 magnification (if a suggestive structure is present) (Fig. 8.3). When the sample is counterstained with chitin-specific Chlorazol Black, fungal structures are accentuated (it takes a dark green or blackish color); this is of particular value if the number of fungal elements is sparse, and it can help to discriminate contaminants such as cotton or elastic fibers, reducing the number of false-positive identifications [3, 23].
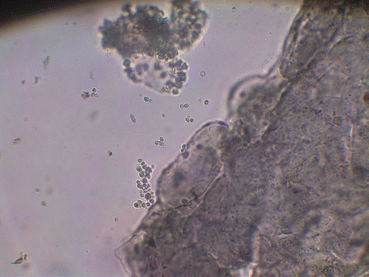

Fig. 8.3
Budding yeasts in direct microscopy (Chlorazol Black, 40X)
It is important to understand the limitations of direct microscopy in diagnosing the cause of onychomycosis, because this technique only serves as a screening test for the presence or absence of fungi, but it could be negative in approximately 15 % of cases, and this technique cannot identify the specific causative agents [3, 23]. Moreover, the sensitivity and specificity of this test are strongly influenced by the clinician’s training. When examining a KOH preparation, the presence of round or ovoid spores suggests yeasts, but pseudohyphae or true hyphae could be present too. Almost half of all specimens taken from onychomycotic nails fail to yield a pathogen in culture. Other diagnostic techniques for a direct examination include Gram, Papanicolaou, Wright, or PAS (periodic acid-Schiff) stains or, in case of its availability, fluorescence microscopy with calcofluor white stain.
In onychomycosis, direct microscopy is the most efficient screening technique; however, careful matching of microscopic and culture results is necessary for the clinician to be confident of the diagnosis to avoid fungal resistance and therapeutic fails [3, 23].
Culture at room temperature (or at 37 ° C) is the only method by which the causative agents can be recognized. Routine culture media such as Sabouraud (with or without chloramphenicol), malt extract, or mycobiotic agar are adequate to obtain the fungal isolates, but cycloheximide inhibits the growth of Candida parapsilosis, C. tropicalis, C. krusei, and C. zeylanoides. The inoculation must be done rapidly because fungal and bacterial contaminants may obscure the real nail pathogen. After 28–72 h at 37 ° C, colonies grow. These organisms produce white, smooth, and brilliant colonies with a creamy consistence. The identification of Candida species is very important to establish the therapeutic regimen. Some chromogenic culture media have been developed to identify the most frequent Candida species. These include CHROMagar®, Biggy® (Fig. 8.4), or Candida-ID® media, and they can identify some species such as Candida albicans, C. tropicalis, C. glabrata, and C. krusei [23].
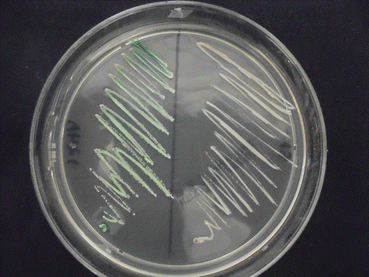

Fig. 8.4
Greenish colony of Candida albicans (CHROMagar® culture medium)
Other no routine laboratory tests to identify different Candida species include serum-induced filamentation (to recognize C. albicans), tetrazolium salt assays, and sensitivity to cycloheximide test.
Also, some systematic biochemical tests are available to identify some species, such as API 20C, API 32C, Vitek®, Uni-Yeast Tek®, Minitek®, Yeast-Ident®, and MicroScan®, which are based on the metabolic and physiologic characteristics of this genus of yeasts [23]. Other identification tools such as ViteK MS®, MALDI TOF/TOF®, and mass spectrometers are used to a wide range of fungi including Candia spp.
Nowadays, molecular diagnostics are a useful tool to specific identification of particular Candida species, and it can be helpful at the moment of deciding the antifungal regime, because some species have an intrinsic drug resistance.
DNA sequencing-based tests are very specific to identify and differentiate among several species of Candida and other microorganisms. In some research centers of China, PCR-based assays combined with internal transcribed spacers sequencing such as ITS1-5.8S-ITS2 rDNA regions are performed to reveal the prevalence of Candida species including emerging species in onychomycosis [24, 25]. Other assay procedures consisted of PCR amplification of the ITS using universal primers, followed by hybridization of the digoxigenin-labeled amplicons to probes on the array [26]. For example, differentiation of C. parapsilosis complex species (Candida parapsilosis sensu stricto, C. metapsilosis, and C. orthopsilosis) is necessary, because C. parapsilosis has shown an intrinsic resistance to fluconazole in vitro. One method to help is the amplification of the secondary alcohol dehydrogenase (SADH) gene and digestion by the restriction enzyme Ban I [27].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree