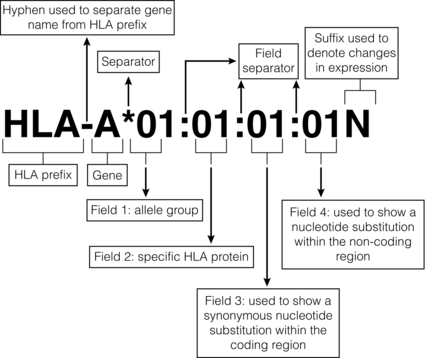
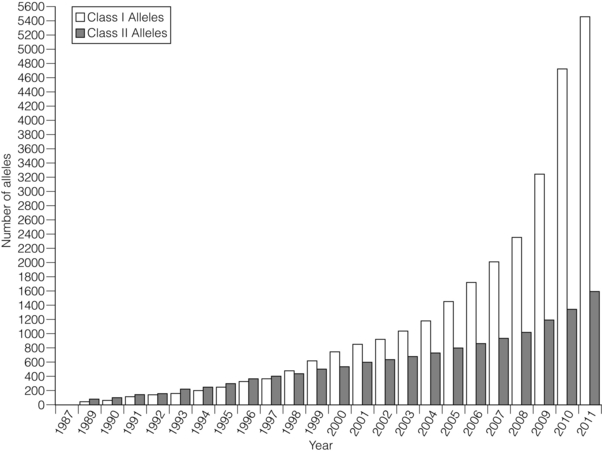
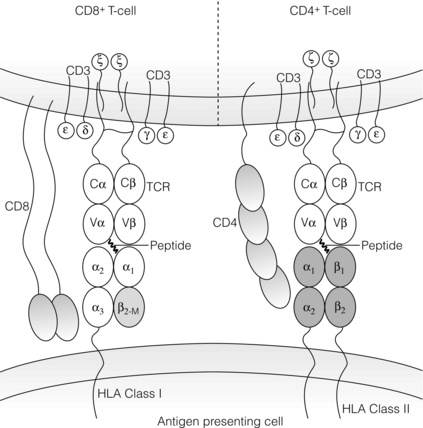
4 ABO-A and -B blood group substances function as histocompatibility antigens as they are expressed as structurally variable carbohydrates, linked to glycoproteins or to glycolipids. The ABO blood group system is unique in that alloantibodies develop naturally in neonates, reaching adult levels within a few years of life, in response to carbohydrates present in the diet. Consequently, ABO-O individuals usually develop antibodies to both ABO-A and -B antigens, ABO-A to -B, ABO-B to -A, but ABO-AB individuals have no ABO blood group antibodies. In the early days of transplantation, pioneers who were aware of the danger of ABO incompatible blood transfusions assumed ABO compatibility should be adopted for organ transplantation. Therefore, the ‘rules’ governing blood transfusion were applied to organ transplantation. It is now known that ABO blood group substances are expressed on endothelium. There has never been a randomised controlled trial of ABO blood group compatible versus incompatible organ transplantation because the risks are perceived to be too high. Of interest is the successful transplantation of ABO blood group incompatible hearts into neonates and the successful transplantation of ABO incompatible kidneys in adults found to have unexpectedly low titre (≤ 1:4) naturally occurring ABO blood group antibodies. Both of these experiences indirectly support the role of ABO blood group substances as histocompatibility antigens in transplantation. In recent years, pre-transplant antibody removal protocols to circumvent the antibody-mediated response to ABO blood group incompatibility in kidney transplantation have been successful.1 To counter the myriad of pathogens that an individual may encounter, the immune system has responded to environmental pressures by selective, adaptive evolutionary processes resulting in the immunogenetic system, which is central to the initial recognition and response to an alloantigen. Human leucocyte antigen (HLA) genes encode highly polymorphic glycoproteins that are expressed on the surface of all nucleated cells, to varying degrees depending on cell function and state of activation. In the natural situation, HLA proteins bind and present self or non-self peptides to T cells, resulting in a respective anergic or vigorous immune response to the antigen presenting cell. Because of their high degree of polymorphism, HLA proteins are also the primary trigger for an alloimmune response (see Chapter 3) and that is why development of clinical transplantation has driven our understanding of HLA gene and allele structure and protein function. Consequently, HLA protein disparity between donor and recipient cells is a barrier to successful allotransplantation, which is only overcome by interventional modulation of the immune response (see Chapter 5). Subsequently, these antibodies can disappear from the circulation, making the identification of prior sensitisation impossible. Infection, which elicits a heightened immune response, can reveal pre-existing HLA reactive alloantibodies because of antigenic epitopes shared between HLA and viral proteins. Furthermore, T cells will be primed to HLA alloantigens. The presence of donor specific HLA reactive IgG antibodies at the time of kidney transplantation conveys a high risk of immediate antibody-mediated hyper-acute rejection (HAR), making the detection and definition of HLA reactive sensitisation an essential component of histocompatibility laboratory services. A crossmatch assay, involving incubation of patient serum with donor target cells, to detect the presence of donor-specific sensitisation is also a prerequisite for successful kidney transplantation in the absence of a virtual crossmatching programme. The culmination of six decades of HLA research is a complete resolution of the major histocompatibility complex of approximately 3.5 megabases of DNA containing at least 25 HLA gene loci coding for in excess of 7000 HLA allele sequences. This is the most polymorphic system in the human genome and exists because of the persistent infections that humans encounter throughout life (Fig. 4.1). Figure 4.1 Polymorphism of the HLA system. Adapted from Robinson J, Mistry K, McWilliam H et al. The IMGT/HLA Database. Nucleic Acids Res 2011; 39(Suppl 1):D1171–6;40 Robinson J, Malik A, Parham P et al. IMGT/HLA – a sequence database for the human major histocompatibility complex. Tissue Antigens 2000; 55:280–7.41 To identify circumstances when HAR is likely to occur, Patel and Terasaki developed the complement-mediated lymphocytotoxicity assay (CDC).3 In outline, this assay comprises a two-stage incubation of donor lymphocytes, as a source of donor HLA proteins, with the potential recipient’s serum and addition of heterologous (rabbit) complement. The CDC assay detects lysis of the target donor lymphocytes through binding of HLA reactive antibody, present in recipient serum, with donor cell surface HLA proteins, mediated by complement-dependent cytotoxicity. When a CDC assay detected recipient serum lysis of donor lymphocytes (a ‘positive’ test), then HAR was shown to be highly likely to occur.4 The second current molecular HLA typing technique employs xMAP® technology, commercially available as the Luminex platform.5 In essence, commercially available kits of Luminex microbeads (5.6 μm), with inherent unique fluorochromasia, are coated with oligonucleotide probes that will hybridise exclusively to the DNA hypervariable region encoding specific HLA alleles. Multiple bead populations, each bearing known HLA-specific oligonucleotide probes, are identified in a multiplex assay by an internal and specific fluorochrome dye and visualised in a flow cytometer with laser light stimulation. Hybridisation to target HLA DNA sequences is revealed by a fluorochrome-tagged streptavidin conjugate that binds to biotin incorporated in the primers used to amplify the HLA locus of interest. There are two types of HLA genes within the human MHC: HLA class I and HLA class II. HLA class I genes encode a single-polypeptide heavy chain that associates with β-2-microglobulin to form an HLA class I protein. HLA class II genes encode both an α (DRA) and a β chain (DRB1, 3, 4, 5), which associate non-covalently to form an HLA class II protein. Both HLA class I and class II proteins are expressed at the cell surface and are bound to a short processed peptide within a peptide-binding site formed by the membrane distal domains of the HLA protein (Fig. 4.2). Figure 4.2 Diagrammatic representation of HLA class I and class II proteins presenting peptide to CD8-positive and CD4-positive T cells. HLA class I and II molecules interact with the T-cell receptor (TCR) complex. The TCR complex consists of the TCR α and β chains; the CD3 γ, δ, and ε chains and 2 ζ chains. Cα and Cβ = Constant region of the α and β TCR chains respectively and Vα and Vβ = Variable region of α and β TCR chains respectively. β2M = β2-microglobulin. Figure prepared and donated by Catherine Wilson. HLA class I proteins are expressed on all nucleated cells and are also present on some non-nucleated cells such as platelets. HLA class II proteins have a restricted cell expression found on the surface of cells involved in antigen presentation, such as dendritic cells and macrophages. However, HLA class II protein expression can be up-regulated on other cell types, including endothelial cells, in response to stimulation by immune mediators such as γ-interferon at the time of immune activation.7 The well-defined WHO nomenclature system is key to the description of HLA allele and protein variation. For description, HLA allele names are broken down into numerical fields that are separated by colons (Fig. 4.3). Not all defined HLA alleles occur at the same frequency. Some combinations of HLA alleles are found on haplotypes at a higher frequency than would be predicted from random association of alleles.11 This observation is called linkage disequilibrium and may result from one or more events, such as:
Testing for histocompatibility
Introduction
Histocompatibility
Sensitisation
HLA: history of clinical application and technical development
HLA genes and proteins: structure and genetics relevant to transplantation
< div class='tao-gold-member'>
![]()
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree