Association studies have recently been revolutionized through newly developed high-throughput SNP genotyping platforms and knowledge gained from the HapMap project, which has shown that the human genome is organized into blocks of haplotypes with limited diversity within each of these blocks, and that as a consequence of this genomic architecture, a limited set of SNPs can capture the vast majority of common variations and ‘tag’ the haplotype pattern sufficiently. Currently available commercial SNP-typing products deploy up to 500,000 to 1,000,000 SNPs. These markers are selected to provide maximal coverage of all common variation via LD-based tagging, or variation within functionally important regions through gene-centric or non-synonymous SNP selections [38]. It has now become routine to carry out hypothesis-free genome-wide surveys to identify common, low-risk variants (i.e. those that are present in more than 5% of the population) that confer a small risk of disease, typically with odds ratios of 1.2–5.0 [39].
Alternative methods to identify genes involved in complex diseases
Alternative approaches to the identification of disease genes in complex diseases include the use of gene expression arrays, and the combination of observed variations in gene expression patterns with genome-wide genotyping to carry out quantitative trait loci (eQTL) mapping. For eczema, so far most reported studies were performed in small sample sizes only and had methodological weaknesses such as inappropriate controls and the use of heterogeneous cellular populations. However, it is anticipated that in the future, well-designed single cell and population cell profiling data from large cohorts and their integration with comprehensive phenotypic data, high-density SNP or resequencing data, and data from proteomic, metabolomic and epigenomic studies as well as from functional screening of genes will greatly enhance the identification and validation of candidate disease genes [40].
Despite their limited applicability to human disease, animal models have aided in identifying genes that affect multifactorial traits [41] and represent a useful complement to human-based genetic studies [42]. The advantages of animal models include a reduced genetic heterogeneity in inbred strains which allows for the dissection of environmental factors in influencing gene regulation under different pathological conditions, the possibility of generating large numbers of offspring in short times and harvesting tissue samples sufficiently, and a higher degree of control over environmental exposure. A number of different murine models of eczema have been reported, but so far there is no reproducible and accessible animal model which combines all the different aspects of eczema [43]. Given the complexity and variability of this disease, it is anticipated that, given the lack of a single comprehensive animal model, many of the disease components will have to be modelled as isolated traits and subsequently combined to explore pathophysiological mechanisms and to provide models for testing new treatments.
References
1 Bieber T. Atopic dermatitis. N Engl J Med 2008;358:1483–94.
2 Wuthrich B. The natural history of atopic dermatitis. In: Ring J, Przybilla B, Ruzicka T (eds) Handbook of Atopic Dermatitis, 2nd edn. Berlin: Springer Verlag, 2006. pp.150–6.
3 Novak N, Bieber T. Allergic and nonallergic forms of atopic diseases. J Allergy Clin Immunol 2003;112:252–62.
4 Wuthrich B, Schmid-Grendelmeier P. The atopic eczema/dermatitis syndrome. Epidemiology, natural course, and immunology of the IgE-associated (“extrinsic”) and the nonallergic (“intrinsic”) AEDS. J Investig Allergol Clin Immunol 2003;13:1–5.
5 Johansson SG, Bieber T, Dahl R et al. Revised nomenclature for allergy for global use: report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol 2004;113:832–6.
6 Spergel JM. Atopic march: link to upper airways. Curr Opin Allergy Clin Immunol 2005;5:17–21.
7 Illi S, von Mutius E, Lau S et al. The natural course of atopic dermatitis from birth to age 7 years and the association with asthma. J Allergy Clin Immunol 2004;113:925–31.
8 Flohr C, Weiland SK, Weinmayr G et al. The role of atopic sensitization in flexural eczema: findings from the International Study of Asthma and Allergies in Childhood Phase Two. J Allergy Clin Immunol 2008;121:141–7.
9 Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immunol 2006;118:209–13.
10 He R, Oyoshi MK, Garibyan L, Kumar L, Ziegler SF, Geha RS. TSLP acts on infiltrating effector T cells to drive allergic skin inflammation. Proc Natl Acad Sci USA 2008;105:11875–80.
11 Demehri S, Morimoto M, Holtzman MJ, Kopan R. Skin-derived TSLP triggers progression from epidermal-barrier defects to asthma. PLoS Biol 2009;7:e1000067.
12 Brenninkmeijer EE, Schram ME, Leeflang MM, Bos JD, Spuls PI. Diagnostic criteria for atopic dermatitis: a systematic review. Br J Dermatol 2008;158:754–65.
13 Johansson SG, Hourihane JO, Bousquet J et al. A revised nomenclature for allergy. An EAACI position statement from the EAACI nomenclature task force. Allergy 2001;56:813–24.
14 Cooke R, van der Veer A. Human sensitization J Immunol 1916;1:201–5.
15 Sneddon IB. The management of infantile eczema. Med Press 1951;226:329–33.
16 Schaffer N. Atopic dermatitis in the older child. J Asthma Res 1966;3:189–91.
17 Schultz Larsen F. Atopic dermatitis: a genetic-epidemiologic study in a population-based twin sample. J Am Acad Dermatol 1993;28:719–23.
18 Larsen FS, Holm NV, Henningsen K. Atopic dermatitis. A genetic-epidemiologic study in a population-based twin sample. J Am Acad Dermatol 1986;15:487–94.
19 Thomsen SF, Ulrik CS, Kyvik KO et al. Importance of genetic factors in the etiology of atopic dermatitis: a twin study. Allergy Asthma Proc 2007;28:535–9.
20 Van Beijsterveldt CE, Boomsma DI. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur Respir J 2007;29:516–21.
21 Phillips PC. Epistasis – the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 2008;9:855–67.
22 Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science 2002;298:2345–9.
23 Williams H, Robertson C, Stewart A et al. Worldwide variations in the prevalence of symptoms of atopic eczema in the International Study of Asthma and Allergies in Childhood. J Allergy Clin Immunol 1999;103:125–38.
24 Asher MI, Montefort S, Bjorksten B et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet 2006;368:733–43.
25 Moffatt MF, Cookson WO. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy 1998;28 Suppl 1:56–61; discussion 5–6.
26 Warner JA, Jones CA, Jones AC, Warner JO. Prenatal origins of allergic disease. J Allergy Clin Immunol 2000;105:S493–8.
27 Diepgen TL, Blettner M. Analysis of familial aggregation of atopic eczema and other atopic diseases by odds ratio regression models. J Invest Dermatol 1996;106:977–81.
28 Dold S, Wjst M, von Mutius E, Reitmeir P, Stiepel E. Genetic risk for asthma, allergic rhinitis, and atopic dermatitis. Arch Dis Child 1992;67:1018–22.
29 Ruiz RG, Kemeny DM, Price JF. Higher risk of infantile atopic dermatitis from maternal atopy than from paternal atopy. Clin Exp Allergy 1992;22:762–6.
30 Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2001;2:21–32.
31 Rice JP, Saccone NL, Corbett J. Model-based methods for linkage analysis. Adv Genet 2008;60:155–73.
32 Almasy L, Blangero J. Contemporary model-free methods for linkage analysis. Adv Genet 2008;60:175–93.
33 McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet 2007;80:588–604.
34 Altmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M. Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet 2001;69:936–50.
35 Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science 2008;322:881–8.
36 Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 2002;3:391–7.
37 Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003;33:177–82.
38 Grant SF, Hakonarson H. Microarray technology and applications in the arena of genome-wide association. Clin Chem 2008;54(7):1116–24.
39 Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet 2009;10:318–29.
40 Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet 2009;10:184–94.
41 Korstanje R, Paigen B. From QTL to gene: the harvest begins. Nat Genet 2002;31:235–6.
42 Hunter KW, Crawford NP. The future of mouse QTL mapping to diagnose disease in mice in the age of whole-genome association studies. Annu Rev Genet 2008;42:131–41.
43 Jin H, He R, Oyoshi M, Geha RS. Animal models of atopic dermatitis. J Invest Dermatol 2009;129:31–40.
Whole-genome linkage studies in eczema
To date, there have been five genome-wide linkage studies performed on eczema. All these whole-genome scans were based on rather small sample sizes and thus statistically significant findings must be interpreted with caution.
The first screen was carried out in 199 German and Scandinavian families using 380 microsatellite markers, and found linkage for eczema and suggestive linkage for total IgE on chromosome 3q21 [1]. The second screen was performed in a panel of 148 British families using 385 microsatellites. This study found evidence of genetic linkage of eczema or eczema plus asthma to three genomic regions, 1q21, 17q25 and 20p. In addition, there was weak evidence for linkage of total serum IgE levels to chromosomes 5q31 and 6qtel [2]. A third genome screen by means of 367 microsatellite markers in 109 Swedish families yielded significant LOD scores for microsatellite markers in chromosome 3p24-p22. For eczema with raised allergen-specific IgE levels, weak evidence for linkage was seen on 18q21. For the semi-quantitative phenotype severity score of eczema in addition to 3q14 suggestive linkage for the chromosomal regions 13q14, 15q14–15 and 17q21 was demonstrated [3]. Using a panel of 446 microsatellite markers, a small-scale Danish study on 23 affected sib-pair families found suggestive linkage to 3p26–3p24, 4p15–4p14 and 18q11–18q12 [4]. In a follow-up study on an independent sample of 130 eczema sib-pair families, the strongest evidence for linkage was obtained for 3p34, 3q21 and 4q22 [5]. A French study on 295 families ascertained through asthmatic probands and genotyped with a panel of 396 microsatellites observed linkage for the combined phenotype asthma+eczema with 5q13 and 11p14. Follow-up fine-mapping of eight markers in the 11p14 locus then suggested a pleiotropic effect for the three allergic diseases eczema, asthma and rhinitis [6].
The only non-European linkage study on eczema investigated 77 Japanese families with 111 affected sib-pairs using a panel of 5861 SNPs instead of microsatellites and found suggestive linkage to 15q21 [7].
The results from these genome screens largely differ from one another, and under a threshold of no more than 10 cM distance between linkage peaks replication can only be considered for the chromosome 3p24 locus [3,4]. The lack of replication of linkage peaks in independent studies might reflect true genetic heterogeneity or be due to differences in ascertainment schemes and phenotype definitions, sample sizes, marker panels and analytical methods between the individual studies. However, the most likely explanation for these inconsistencies might be the limitations of linkage studies for the dissection of complex traits, especially when utilized on small sample sizes. Apart from FLG, which accounts for part of the significant linkage signal on 1q21 [8], the underlying disease genes remain elusive. Interestingly, the regions linked to eczema show only very limited overlap with those known for asthma, suggesting that susceptibility to asthma and eczema is mediated through different genes rather than through a shared susceptibility to a common atopic background. The AE loci identified thus far are, however, closely coincident with psoriasis susceptibility regions, e.g. 1q21, suggesting that these conditions share susceptibility loci in genomic regions encoding tissue-specific gene, perhaps with general effects on dermal inflammation, immunity and structure and an important role for proteins expressed by epithelial cells [9].
References
1 Lee YA, Wahn U, Kehrt R et al. A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nat Genet 2000;26:470–3.
2 Cookson WO, Ubhi B, Lawrence R et al. Genetic linkage of childhood atopic dermatitis to psoriasis susceptibility loci. Nat Genet 2001;27:372–3.
3 Bradley M, Soderhall C, Luthman H, Wahlgren CF, Kockum I, Nordenskjold M. Susceptibility loci for atopic dermatitis on chromosomes 3, 13, 15, 17 and 18 in a Swedish population. Hum Mol Genet 2002;11:1539–48.
4 Haagerup A, Bjerke T, Schiotz PO et al. Atopic dermatitis – a total genome scan for susceptibility genes. Acta Derm Venereol 2004;84:346–52.
5 Christensen U, Moller-Larsen S, Nyegaard M et al. Linkage of atopic dermatitis to chromosomes 4q22, 3p24 and 3q21. Hum Genet 2009;126(4):549–57.
6 Guilloud-Bataille M, Bouzigon E, Annesi-Maesano I et al. Evidence for linkage of a new region (11p14) to eczema and allergic diseases. Hum Genet 2008;122:605–14.
7 Enomoto H, Noguchi E, Iijima S et al. Single nucleotide polymorphism-based genome-wide linkage analysis in Japanese atopic dermatitis families. BMC Dermatol 2007;7:5.
8 Morar N, Cookson WO, Harper JI, Moffatt MF. Filaggrin mutations in children with severe atopic dermatitis. J Invest Dermatol 2007;127:1667–72.
9 Willis-Owen SA, Morar N, Willis-Owen CA. Atopic dermatitis: insights from linkage overlap and disease co-morbidity. Expert Rev Mol Med 2007;9:1–13.
Whole-genome association studies in eczema
As outlined above, simultaneous examination of up to 1,000,000 SNPs in a single high-throughput assay can now be efficiently used to accurately capture (or ‘tag’) the vast majority of the diversity in the genome and search for loci associated with a disease [1]. Different SNP arrays with varying SNP content and density, which were designed using diverse marker selection strategies, are now commercially available, and their power to interrogate a significant proportion of human genetic variation has facilitated hundreds of genome-wide association studies (GWAS) and has led to the identification of numerous novel susceptibility loci for many complex traits [2].
The first and to date only GWA screen of eczema, published in 2009, incorporated more than 307,000 SNP markers and 10,000 individuals [3]. The cohort comprised family and case-referent panels mainly collected from Germany, allowing internal replication of results. This first GWAS in eczema identified a robust site of association on chromosome 11q13.5, with the strongest association observed for SNP rs7927894, which is located in an intergenic region between two annotated genes: chromosome 11 open reading frame 30 (C11orf30) and leucine-rich repeat containing 32 (LRRC32) (UCSC Genome Browser, Human Genome March 2006 assembly, 259: http://genome.ucsc.edu). This association has recently been confirmed in another independent population of Irish paediatric eczema cases [4]. Interestingly, the same risk allele (rs7927894 allele A) also showed an association with Crohn disease in an independent GWAS [5]. The causative gene or gene product defined by this SNP, however, remains to be identified and functionally characterized. C11orf30 encodes the nuclear protein EMSY, which binds and inactivates the cancer susceptibility gene BRCA2, and has been implicated in chromatin modification, DNA repair and transcriptional regulation. In addition, an increase in C11orf30 copy number has been reported in epithelium-derived cancer of the breast and ovary. The potential involvement of C11orf30 in multiple inflammatory and malignant epithelial diseases suggests a role for C11orf30 in epithelial immunity, growth, and/or differentiation.
The second nearby gene, LRRC32 (also known as GARP), has recently been shown to be a cell surface molecule expressed on regulatory T-cells, and therefore also represents an attractive candidate for eczema susceptibility. However, it is also possible that rs7927894 is in LD with causative variants in more distant genes. Clearly, further replication in additional cohorts, fine-mapping and functional studies will be required to identify the causative variant or variants at this locus. The same study reported an additional susceptibility SNP located within the HRNR gene, which encodes the filaggrin-related protein hornerin, on chromosome 1q21, but so far there is no independent confirmation of this finding. In addition, the association of four prevalent FLG null mutations with eczema could again be confirmed, and putative, nominally significant associations of SNPs in six additional loci were observed. Some of these loci affect immune processes related to eczema pathogenesis such as the differentiation of type 2 T helper cells, mast cell, eosinophil and dendritic cell chemotaxis, and pruritus (HRH4), toll-like receptor-mediated responses to bacteria (LY86, also called MD-1), and CD8+ T-cell function (EOMES) and thus represent candidate genes of potential interest.
Genome-wide association studies have also resulted in the identification of novel susceptibility genes for asthma (ORMDL3, CHI3L1, PDE4D) [6–8], as well as variants in the gene encoding the high affinity receptor for IgE (FCER1A) for total IgE [9]. These genes have been replicated and functional studies have supported their relevance in asthma and atopy, whereas their potential role in eczema is not yet clear.
In general, GWA studies have already identified a large number of robust associations between specific chromosomal loci and complex human diseases (yet explaining only a modest proportion of trait heritability) and clearly represent a powerful new tool for the genetic dissection of complex diseases, which, however, needs to be kept in perspective. GWA approaches present many logistical, technical and biostatistical challenges, and have several significant limitations. These limitations include false-positive and false-negative results and susceptibility to bias through genetic and disease heterogeneity, the need for large sample sizes, the insensitivity to rare variants (such as FLG mutations), structural variants (such as copy-number variants), and variants in recombinational hotspots, the need for follow-up investigations (to detect causal variants) and the lack of information on gene function [2]. However, GWA studies clearly have the potential to significantly advance our understanding of the genetics and pathogenetic mechanisms of complex diseases such as eczema, if the studies are performed under stringent conditions, are sufficiently powered, and are thoroughly reproduced [10].
References
1 McCarthy MI, Abecasis GR, Cardon LR et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–69.
2 Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet 2009;10:241–51.
3 Esparza-Gordillo J, Weidinger S, Folster-Holst R et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet 2009;41:596–601.
4 O’Regan GM, Campbell LE, Cordell HJ, Irvine AD, McLean WH, Brown SJ. Chromosome 11q13.5 variant associated with childhood eczema: an effect supplementary to filaggrin mutations. J Allergy Clin Immunol 2010;125:170–4.
5 Barrett JC, Hansoul S, Nicolae DL et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008;40:955–62.
6 Moffatt MF, Kabesch M, Liang L et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–3.
7 Ober C, Tan Z, Sun Y, Possick JD et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med 2008;358:1682–91.
8 Himes BE, Hunninghake GM, Baurley JW et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet 2009;84:581–93.
9 Weidinger S, Gieger C, Rodriguez E et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet 2008;4:e1000166.
10 Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med 2009;360:1759–68.
Candidate gene studies in eczema
Until recently, association studies in eczema have focused on candidate genes, pathways or chromosomal regions. Prior to the detection of filaggrin (FLG, see below) and based on the assumption that the primary defect is immunological, mostly candidate genes encoding major elements of the immune system were investigated. In a search of public databases up to June 2009, a recent review has identified more than 100 hypothesis-driven studies which reported results of tests for association for eczema on a candidate gene, and considering publication bias there are undoubtedly more genes for which associations have been tested but failed [1]. However, so far candidate gene studies mostly yielded inconsistent results. Only very few associations could be replicated in at least one independent study (see Table 23.1) and the specific phenotypes found to be associated varied, possibly indicating a role for the atopic state rather than eczema per se. Furthermore, apart from FLG, even for replicated genes a considerable number of negative reports exist. Candidate genes for which the balance of evidence weighs in favour of a role in eczema pathogenesis are presented and discussed in more detail below. These genes can be considered as falling into two major functional groups: genes encoding epidermal or other epithelial structural proteins, and genes contributing to immune dysregulation.
Table 23.1 List of the genes and their chromosomal location that have been associated with eczema in at least two independent study samples
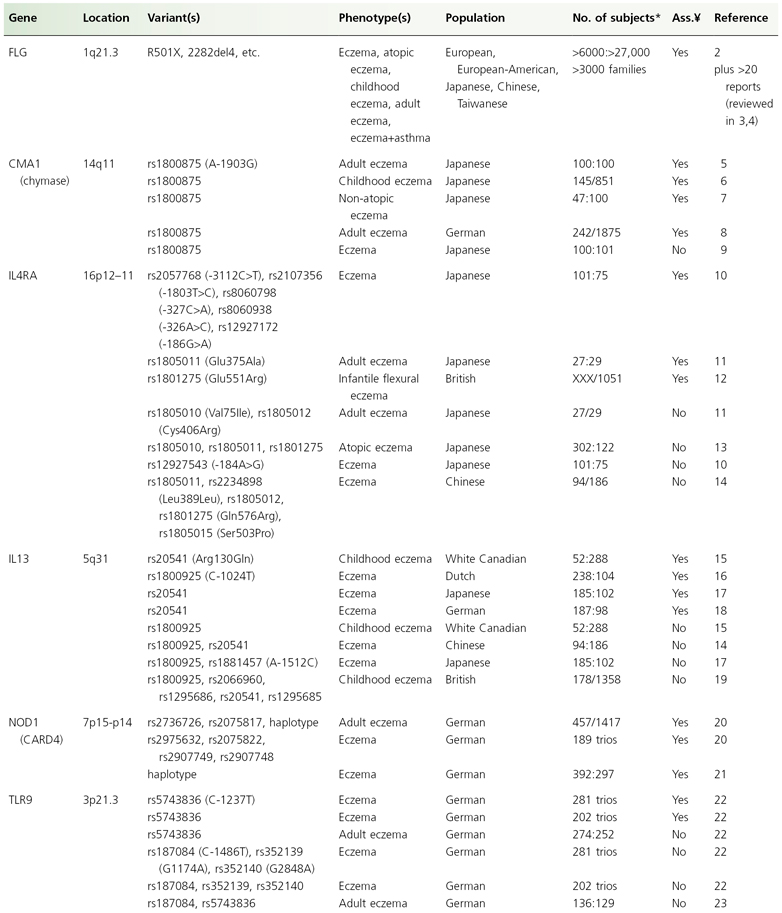
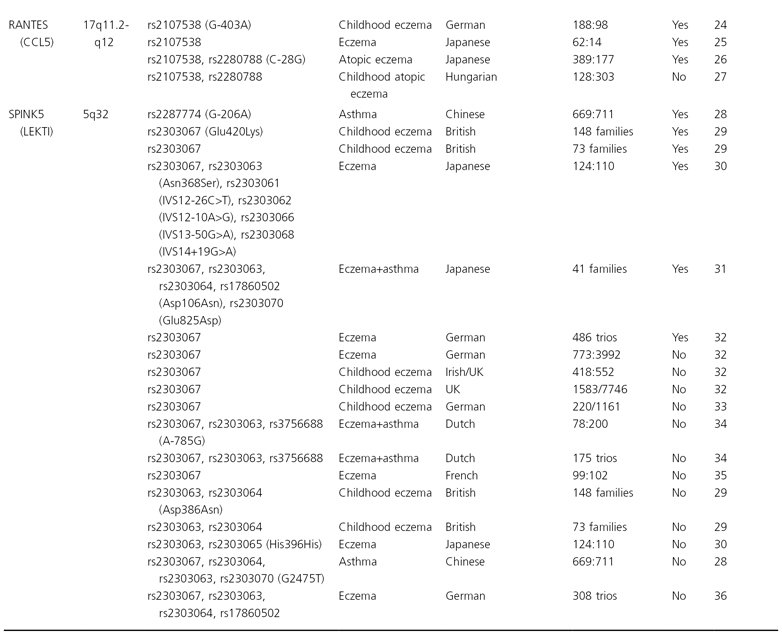
* Cases:controls, cases/population; ¥, association.
Genes implicated in epithelial barrier dysfunction
Filaggrin (FLG)
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








