Tumor type
Translocation/chromosome involved
Genes involved
Alveolar rhabdomyosarcoma
t(2;13))(q35;q14)
t(1;13)(p36;q14)
t(X;2)(q13;q35)
PAX3-FKHR
PAX7-FKHR
PAX3-AFX
Alveolar soft-part sarcoma
der(17)t(X;17)(p11;q25)
ASPL-TFE3
Angiomatoid fibrous histiocytoma
t(12;22)(q13;q12)
t(2;22)(q33;q12)
t(12;16)(q13;p11)
EWSR1-ATF1
EWSR1-CREB1
FUS-ATF1
Chondroid lipoma
t(11;16)(q13;p13)
CLLorf95-MKL2
Clear-cell sarcoma
t(12;22)(q13;q12)
t(2;22)(q33;q12)
EWSR1-AFT1
EWSR1-CREB1
Congenital/infantile fibrosarcoma
t(12;15)(p13;q25)
ETV6-NTRK3
Dermatofibrosarcoma protuberans (DFSP)
t(17;22)(q21;q13)
COLIA1-PDGFB
Desmoid fibromatosis
3p21 5q22
CTNNB1 (sporadic) APC (FAP related)
Desmoplastic small round cell tumor
t(11;22)(p13;q12)
EWSR1-WT1
Epithelioid hemangioendothelioma
t(1;3)(p36;q25)
t(11;21)(q22;q22)
WWTR1-CAMTA1
YAP1-TFE3
Epithelioid sarcoma
22q11.23
SMARCB1
Ewing sarcoma: PNET
t(11;12)(q24;q12)
t(21;22)(q22;q12)
t(7;22)(p22;q12)
t(2;22)(q33;q12)
t(17;22)(q12;q12)
inv(22)(q12;q12)
t(16;21)(p11;q22)
EWSR1-FLI1
EWSR1-ERG
EWSR1-ETV1
EWSR1-FEV
EWSR1-E1AF
EWSR1-ZSG
FUS-ERG
“Ewing-like” tumors
t(4;19)(q35;q13)
inv(X)(p11.4;p11.22)
CIC-DUX4
BCOR-CCNB3
Extraskeletal myxoid chondrosarcoma
t(9;22)(q22;q12)
t(9;17)(q22;q11)
t(9;15)(q22;q21)
t(3;9)(q11;q22)
EWSR1-NR4A3
TAF2N-NR4A3
TCF12-NR4A3
TFG-NR4A3
Gastrointestinal stromal tumor
4q12 4q12
KIT PDGFRFA
Inflammatory myofibroblastic tumor
t(1;2)(q22;p23)
t(2;19)(p23;p13)
t(2;17)(p23;q23)
t(2;2)(p23;q13)
t(2;11)(p23;p15)
inv(2)(p23;q35)
TPM3-ALK
TPM4-ALK
CLTC-ALK
RANB2-ALK
CARS-ALK
ATIC-ALK
Lipoblastoma
8q11-13
PLAG1, HMGA2 with various partners
Lipoma
12q13 ~ 15, 13q, 6p21 ~ 23
HMGA2 with various partners (LPP, CXCR7, EBF1, NFIB, LHFP), subset of tumors
Low-grade fibromyxoid sarcoma
t(7;16)(q33;p11)
t(11;16)(p11;p11)
FUS-CREB3L2
FUS-CREB3L1
Mesenchymal chondrosarcoma
t(8;8)(q13;q21)
HEY1-NCOA2
Myoepithelial tumor of soft tissue
t(6;22)(p21;q12)
t(19;22)(q13;q12)
t(1;22)(q23;q12)
t(1;16)(p34;p11)
t(1;22)(p34;q12)
8q12 rearrangements
EWSR1-POU5F1
EWSR1-ZNF444
EWSR1-PBX1
FUS-KLF17
EWSR1-KLF17
PLAG1
Myxoid/round-cell liposarcoma
t(12;16)(q13;p11)
t(12;22)(q13;q12)
FUS-DD1T3(TLS-CHOP)
EWSR1-DD1T3(EWSR1-CHOP)
Myxoinflammatory fibroblastic sarcoma/hemosiderotic fibrolipomatous tumor
t(1;10)(p22;q24)
3p11 ~ 12(ring chromosome)
TGFBR3 ~ MGEA5, Amplification of VGLL3, CHMP2B
Nodular fasciitis
t(17;22)(p13;q13)
MYH9-USP6
Pseudomyogenic hemangioendothelioma
t(7;19)(q22;q13)
SERPINE1-FOSB
Sclerosing epithelioid fibrosarcoma
t(7;16)(p22;q24)
FUS-CREB3L2
Soft tissue angiofibroma
t(5;18)(p15;q13)
AHRR-NCOA2
Solitary fibrous tumor
inv(12)(q13;q13)
NAB2-STAT6
Synovial sarcoma
t(X;18)(p11;q11)
SS18-SSX1, SSX2, SSX4
Tenosynovial giant cell tumor/pigmented villonodular synovitis
t(1;2)(p13;q37)
COL6A3-CSF
Well differentiated liposarcoma/ALT/dedifferentiated liposarcoma
12q13 ~ 15(ring chromosomes, giant marker chromosomes)
Amplification of MDM2, CDK4, HMGA2, GLI, SAS, others
Previously, particular chromosomal rearrangements were considered to be specific to a particular mesenchymal tumor. However, this concept has evolved over several years as it is now understood that multiple tumors can have identical rearrangements (see Fig. 3.1). Chimeric genes involving EWSR1 or FUS demonstrate this point well. Both genes are prominent players with many tumors and in some instances seem to even be used interchangeable as oncogenic promoters by the same tumor [1, 5, 14, 15]. These same genes are also not exclusive to mesenchymal tumors but can be seen in tumors of other lineages, including leukemia and carcinoma. Not only can mesenchymal tumors involve the same gene but they can share the exact same translocations. The most commonly cited example of this is clear cell sarcoma and angiomatoid fibrous histiocytoma [16–20]. Both have translocations involving EWSR1 (22q12) and ATF1 (12q13) and/or CREB1 (2q34), yet are phenotypically divergent. Clear cell sarcoma is an aggressive tumor with melanocytic differentiation, while angiomatoid fibrous histiocytoma is an indolent tumor with uncertain differentiation. Although important to tumoriogenesis, other factors are clearly involved for phenotypic development in these tumors besides the single translocation. Interestingly, some of these same translocations can be seen in another recently described sarcoma, primary pulmonary myxoid sarcoma, and also carcinomas, such as hyalinizing clear cell carcinoma of the salivary gland and clear cell odontogenic carcinoma [21–24]. Even amplification involving the same regions can be seen in very different tumors, such as well-differentiated liposarcoma and well-differentiated osteosarcoma, both of which have amplification of 12q13 ~ 15 [25]. This phenomenon of shared genes, translocations and genetic alterations also highlights the importance of correlating molecular results with histology, immunohistochemical, and clinical findings to ensure that the correct interpretation is made. With advances in next generation sequencing techniques, additional discoveries continue to further our understanding of these rare tumors which can be useful diagnostically, as well as offer a potential therapeutic target.
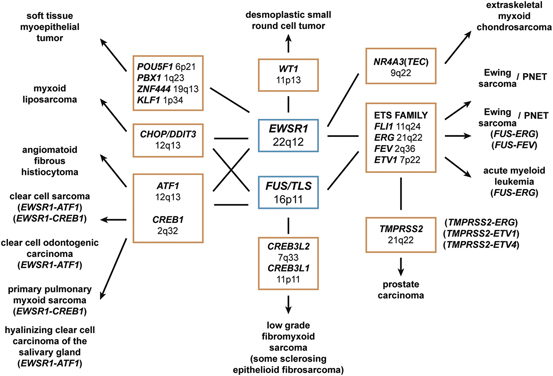

Fig. 3.1
Multiple mesenchymal tumors can share rearrangements in the same gene and even identical fusion transcripts.
Molecular Techniques
Multiple ancillary molecular techniques are available to pathologists. Each technique carries unique strengths and weaknesses. It is important to be familiar with these features as they may influence which assay is best for a given situation. Cytogenetic karyotyping has been long considered the gold standard in detecting genetic aberrations , and has given rise to many of the discoveries today [6]. Fresh tissue is obtained so that tumor cells can be grown and then processed so that metaphase chromosomes can be examined (Fig. 3.2). Fluorescently labeled probes can sometimes be applied to identify the chromosomes (also known as spectral karyotyping) or to detect rearrangements of specific genes (see below for fluorescence in situ hybridization). An advantage of this technique is that it requires no prior knowledge of the genetic aberration and thus can detect a wide range of translocations and chromosomal abnormalities. This is particularly useful when dealing with difficult-to-diagnose mesenchymal tumors and where the identification of an unexpected translocation may provide clues to further classification, such as unusual histological or immunohistochemical variants. However, this technique requires fresh tissue, which is not always available. Lengthy processing time, expert technical staff, complex interpretation, bacterial contamination or normal tissue overgrowth, and associated costs can be negative issues. Finally, the technique cannot detect small mutations and translocations where the genes are in close proximity to each other (also known as cryptic translocations.) As a result, conventional karyotyping has largely given way to faster molecular techniques which have been optimized for formalin-fixed paraffin-embedded (FFPE) tissue .


Fig. 3.2
Karyotype of a case of clear cell sarcoma showing a characteristic translocation involving chromosomes 12 and 22.
One of the most widely used molecular technique for detecting translocations is fluorescence in situ hybridization (FISH) because of its robustness and broad utility in frozen, FFPE and cytological tissues. In this technique, fluorescently labelled DNA probes are applied to unstained slides. Interphase nuclei are examined under fluorescence microscope [6]. The most commonly available FISH assays are break-apart assays. In break-apart assays, two DNA probes flank a single gene of interest (Fig. 3.3). For example, in the EWSR1 break-apart assay, two DNA probes flank the EWSR1, one in the 5ʹ end labeled with a probe which will fluorescence orange and one in the 3ʹ end with a probe which will fluorescence green. In tumors where EWSR1 is not rearranged, both orange- and green-labeled probes co-localize to produce a yellow signal because of their spectral overlap when in close proximity to each other. In contrast, tumors where EWSR1 is rearranged will have split signals, one separate orange and one separate green signal. FISH assays with fusion probes will also have two DNA probes but one for every involved genes. For example, in a fusion FISH assay for Ewing sarcoma, one probe will be bound to the 5ʹ EWSR1 end, while a second probe will be bound to the 3ʹ end of FLI1. In contrast to the break-apart assays, tumors without rearrangement will have split signals, while tumors with rearrangements will have both probes next to other, co-localizing into a single yellow signal. In practice, break-apart FISH is a much more common approach as it covers a wider range of events with a single set of probes. The strengths of using FISH include its robustness in multiple tissues; most notably in FFPE tissue, the most commonly available tissue. Because it is in situ, the test is interpreted within tumor nuclei, something not possible in polymerase chain reactions (PCRs; see below). Furthermore, it is much faster than karyotyping as it does not require time for cells to grow. One disadvantage of FISH is that prior knowledge of a gene to target is needed. Furthermore, most commercially available FISH are break-apart assays that do not provide information on partner. Therefore, it cannot be used to differentiate similar appearing entities which have the same gene involved. For example, Ewing sarcoma and desmoplastic small round cell sarcoma are both small round cell tumors and both have rearrangements involving EWSR1, though with different partners. Break-apart FISH for EWSR1 cannot be used to distinguish between these two entities. Another disadvantage is that multiplexing (ability to screen multiple gene aberrations at the same time) is limited as each assay typically requires an unstained slide, and is compounded by the fact that only a few DNA probes are commercially available .
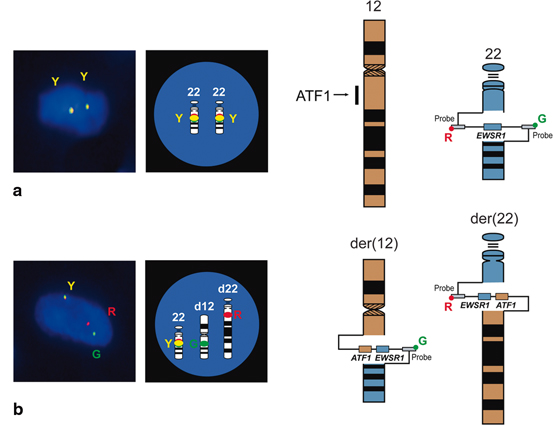

Fig. 3.3
Break-apart fluorescence in situ hybridization (FISH) assay for EWSR1. Example shown is of clear cell sarcoma. a. In tumors which are negative for rearrangement, DNA probes flanking EWSR1 (one green and one red) co-localize to produce a yellow signal. b. In tumors with EWSR1 rearrangement, the signals are split.
Polymerase chain reaction (PCR) and reverse transcriptase-polymerase chain reaction (RT-PCR) have become common ancillary diagnostic assays for mesenchymal tumors [6]. DNA primers flank the gene or fusion gene of interest, are amplified several fold and sequenced to determine if mutation is present or not (Fig. 3.4). In RT-PCR , fusion transcript RNA is converted to DNA (cDNA) first. The major advantage of this technique is that it detects a specific gene fusion, providing information on both gene partners. This is important when knowledge of partner genes is needed to differentiate between two similar appearing tumors. As mentioned earlier, break-apart FISH for EWSR1 would not be useful to distinguish between Ewing sarcoma and desmoplastic small round cell tumor. However, RT-PCR would be able to discriminate between Ewing sarcoma (EWSR1-FLI1 or EWSR1-ERG) and desmoplastic small round cell tumor (EWSR1-WT1), because it can specify fusion partners. Also, RNA extracted from unstained slides can be used to do multiple reactions. The major disadvantage of this technique is that it is not as robust as FISH. RNA is susceptible to degradation particularly in older FFPE blocks and decalcification. In addition, the majority of laboratories have assays which only detect the most common fusion transcript types and variants. A fusion transcript type is created between variations in breakpoints between the same genes, each of which is designated numerically as a type. For example, in Ewing sarcoma, there are multiple EWSR1-FLI1 fusion types, with types I and II being most common. A fusion transcript variant is created with different genes. In Ewing sarcoma, EWSR1 is most commonly paired with FLI1; however, it can also be paired with ERG, FTV1, and FEV among others to yield other variants. Therefore, it is possible that a less common variant or type is missed because the assay is not designed to detect them. One particular problematic tumor is dermatofibrosarcoma protuberans (DFSP) because it has multiple types due to substantial variation in break points within COL1A1 which comprise more than 50 exons. Therefore, multiple primers are needed to cover these many types [26]. Similar to FISH, commercial laboratories only offer a selected few RT-PCR assays. Finally, lack of in situ testing, raises the possibility that contamination can occur. Therefore, it is important to be confident that the laboratory chosen for such studies has solid protocols and impeccable procedures .
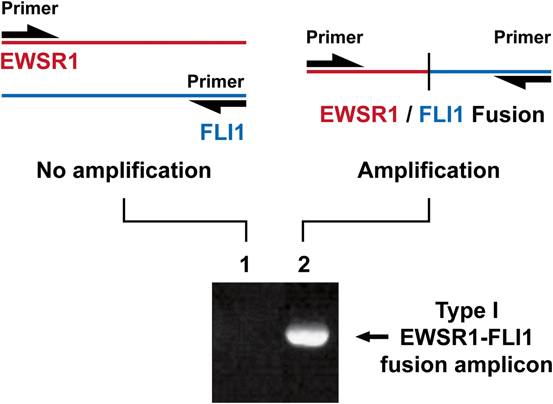

Fig. 3.4
PCR primers flank DNA or cDNA of interest (example shown is EWSR1-FLI1 cDNA). Samples containing a fusion transcript will result in a band on electrophoresis which can be sequenced for confirmation.
An exciting development in molecular diagnostics is the increasing utilization of next generation sequencing techniques. While global RNA sequencing (RNAseq) is not ready for broad clinical application, semi-anchored approaches are being explored where primers specific for a common known gene is used with opposing nonspecific primers to theoretically identify any partner gene (Fig. 3.5). The bioinformatics for interpreting fusions is rapidly evolving and this is a growth area in molecular testing for translocations. Next generation sequencing can also be used to examine for broad panels of genes for point mutations, insertions, and deletions (and increasingly copy number alterations). These approaches have limited applications in sarcoma as the genomic properties of most sarcomas are poorly examined, but this is rapidly changing as new genomic analyses of sarcomas are being published regularly.
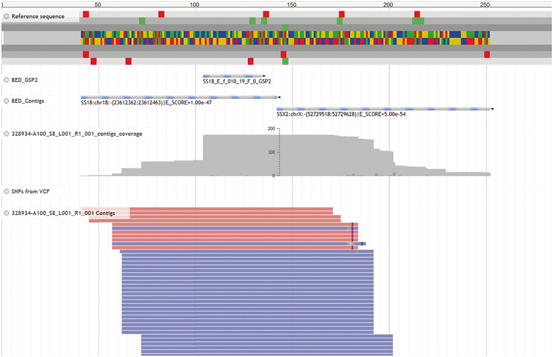

Fig. 3.5
While still evolving in both technique and bioinformatics interpretation, next generation sequencing approaches using semi-anchored amplification techniques are beginning to be employed in the clinical setting.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








