4 Testing for histocompatibility
Introduction
The immune system has evolved specifically to recognise and destroy hazardous infective agents such as bacteria and viruses. These same mechanisms function to reject non-self-allogenic tissues, which are an irritant to surgical transplantation. Successful organ transplants occur between monozygotic twins (syngeneic transplants) or between genetically related or unrelated individuals (allogeneic transplants) – but only when the immune system is hindered by effective immunosuppression. The degree of immunosuppression needed, which reflects the frequency and strength of the alloimmune response, will be determined by the immunogenetic disparity between the donor and the recipient. The ability of a recipient to respond to allogeneic tissue will reflect their own immunogenetic constitution – their immune responsiveness. There are two major genetic systems determining human allogenicity, each of which can convey a biological veto or a biological impediment to effective clinical transplantation.
In the situation of biological impediment, effective clinical transplantation can only be achieved through use of special protocols to remove circulating antibody (see Chapter 7).
HLA genetics
The human leucocyte antigen (HLA) complex is located on the short arm of chromosome 6 at 6p21.3. This region is also known as the major histocompatibility complex (MHC). This collection of highly polymorphic genes codes for HLA molecules, which are cell-surface proteins that play a pivotal role in antigen presentation and recognition, and hence the survival of transplanted organs and tissues. For the purposes of this chapter the emphasis will be placed on elements of the HLA system that are characterised on a routine basis in the histocompatibility and immunogenetics laboratory.
There are two classes of HLA genes that are involved in the immune response to transplanted organs, class I and class II. These classes are structurally and functionally distinct. Class I molecules are involved in the processing and presentation of intracellular peptide to CD8+ T cells. Class II molecules process and present extracellular peptide to CD4+ T cells. This process is described in more detail in Chapter 3 on transplantation immunology.
HLA nomenclature
Nomenclature for both accepted and novel HLA alleles is regulated by the World Health Organisation (WHO) Nomenclature Committee for factors of the HLA system.1 HLA sequences are officially recorded on the IMGT/HLA Sequence Database (www.ebi.ac.uk/imgt), which is updated quarterly and is part of the International Immunogenetics Project (IMGT). HLA genes are highly polymorphic. In January 2008, this database contained sequences for 2047 class I alleles and 944 class II alleles.
Over time, major revisions of HLA nomenclature are necessary due to the ever-increasing numbers of alleles identified. A guide to the most recent nomenclature for HLA antigens and alleles is summarised in Table 4.1, where resolution of HLA alleles to the four-digit level is shown. In practice matching beyond this level, even in stem cell transplantation, is impractical. Resolution to eight digits can be carried out and in some instances an alphabetical suffix is used to describe the biological expression of an encoded molecule.
Table 4.1 HLA nomenclature: basic overview of the levels of HLA typing performed in the histocompatibility laboratory
| WHO nomenclature | Interpretation |
|---|---|
| HLA-B | Identification of HLA locus |
| HLA-B44 | HLA defined by serology-based technique |
| HLA-B*44 | Asterisk denotes HLA alleles defined by analysis of DNA |
| HLA-B*44 two-digit resolution | Denotes the allele family |
| Corresponds where possible to the serological group | |
| Often termed ‘low resolution’ | |
| Level used for matching in solid organ transplants | |
| HLA-B*4402 four-digit resolution | Allele sequence variation results in amino acid substitutions, coding variation or non-synonymous changes |
| Level of matching used in haemopoietic stem cell transplantation |
HLA typing
Molecular techniques
Molecular techniques for HLA typing of DNA sequence polymorphisms have largely replaced serology since they offer flexibility of resolution, much improved reproducibility and greater accuracy. The invention of the polymerase chain reaction (PCR)2 has revolutionised HLA typing techniques by facilitating identification of HLA polymorphisms at the single nucleotide polymorphism (SNP) level.
Principle of the polymerase chain reaction
This process is performed in a thermocycler, which creates rapid (millisecond) changes in temperature in a controlled environment.
PCR sequence-specific primers (PCR-SSP)
PCR-SSP is currently the HLA typing system of choice in most histocompatibility and immunogenetics laboratories for typing deceased organ donors. A result can be generated in 3 hours. There are various commercially available PCR-SSP kits in use, such as those manufactured by Invitrogen (www.invitrogen.com), Olerup (www.alphahelix.co.uk) and BAG Health Care GmbH (www.bag-germany.com). These kits are constantly updated by the manufacturer to incorporate new WHO-recognised alleles. Some laboratories construct in-house SSP trays using their own design primers; however, as more alleles are defined it has become almost impossible for individual laboratories to keep their own primer design up to date.
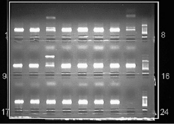
The assignment of alleles is based on the presence or absence of amplified product. To ensure that the absence of a specific product is due to the individual lacking the corresponding sequence and is not simply due to a technical error, a control is included in each of the SSP reactions. The control is an invariant region of a gene that is constant between all individuals, such as human growth hormone. The PCR-SSP product is visualised by size differences using agarose gel electrophoresis, as shown in Fig. 4.1. Electrophoresis through agarose relies on the movement of negatively charged DNA (due to the phosphate backbone) towards the anode. Fragments of DNA differentially migrate and thus can be identified according to their size. DNA is visualised on the gel by staining with ethidium bromide, which intercollates between the strands of DNA and fluoresces under ultraviolet light. In Fig. 4.1, control bands are visible in each of the SSP reactions and the specific product is visible as a second band in the well. By knowing which wells contain which SSP, the HLA type of an individual can be allocated.
PCR sequence-specific oligonucleotide probes (PCR-SSOP)
PCR-SSOP was the first PCR-based technique used for detecting HLA polymorphisms.3,4 The technique has advantages over PCR-SSP, in particular a large sample throughput can be achieved. Until recently the methodology and interpretation of results was complex and time-consuming, which meant that it was not suitable for deceased donor HLA typing.
Microbead array technology
Further details, along with a step-by-step animation of the principles of this technology, can be viewed at www.luminex.corp.
RSSO in conjunction with microbead array technology
Target DNA is amplified in the PCR using primers that are labelled with biotin. The double-stranded DNA is then denatured to leave single-stranded DNA. At this point SSO probes bound to fluorescent microspheres are added. The probes determine the specificity of the reaction and different numbers of probes are used depending on the locus to be tested. The SSO probes bind to allele-specific regions of DNA. When the beads are washed any unbound probes will be removed. The addition of PE-conjugated streptavidin then causes a chemical reaction with biotin to release fluorescence, which is detected by a dedicated analyser (e.g. LABscan™ 100). The analyser in conjunction with an XY platform and sheath delivery system means that 96-well trays can be read automatically. Software is then used to analyse the fluorescence pattern (probes bound) and matches them to the patterns of known alleles, hence defining the HLA type. Several manufacturers provide RSSO kits and technology, including LABType™ produced by OneLambda (www.onelambda.com) and Lifematch™ produced by Tepnel Life Sciences.
Sequence-based typing (SBT)
SBT involves typing of HLA genes at the nucleotide or allele level. The technique utilises the elegant way in which DNA is copied by a DNA polymerase enzyme. Bases are incorporated into an extending DNA strand in a PCR through the formation of a phosphodiester bridge between the 3′ hydroxyl group at the growing end of a primer and the 5′ phosphate group of the nucleotide that is being incorporated.5 In SBT, DNA polymerases incorporate analogues of A, G, T and C that do not have a 3′ hydroxyl group. This results in termination of the strand.6 The analogues of A, G, T and C are labelled with a fluorescent dye. Detection of the dye then reveals which base is incorporated at each position. In this way the entire sequence can be determined. There are various SBT techniques and software options available from manufacturers such as Abbott Diagnostics (www.abbottmolecular.com) and Protrans (www.protrans.com) for HLA typing.
Causes of HLA-specific alloimmunisation
 HLA-specific antibodies can be produced in response to blood transfusions, pregnancy or transplantation.7–9
HLA-specific antibodies can be produced in response to blood transfusions, pregnancy or transplantation.7–9
Blood transfusion
The antigen source in a blood transfusion is the lymphocyte content of the transfused blood.10 A number of studies have indicated that a low number of transfusions are a minimal risk for sensitisation whilst multiple transfusions are required to cause alloimmunisation.11,12 These data suggest that one or a few – usually less than 10 – blood transfusions induce minimal clonal expansion with limited proliferation of T and B cells. This is usually a transient IgM antibody, or IgM antibody followed by IgG antibody production, which is promptly downregulated. Multiple transfusions induce significant clonal expansion where antibody production is more likely and may result in a sustained level of IgG antibody production.13
Pregnancy
HLA-specific antibodies can be produced during pregnancy as a result of stimulation by paternal HLA antigens expressed on the fetus. Stimulation occurs when high levels of antigen enter maternal circulation during the birth of the first child. Subsequent pregnancies may induce a secondary response. Multiparous female patients are at greater risk of sensitisation following blood transfusions, suggesting that a degree of clonal expansion has occurred during pregnancy so that antibodies are more easily induced by subsequent blood transfusions.14
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree


 HLA typing and evaluation of HLA mismatching are essential prerequisites to kidney and stem cell transplantation.
HLA typing and evaluation of HLA mismatching are essential prerequisites to kidney and stem cell transplantation. Available evidence suggests that the immune system of all allograft recipients undergoes clonal expansion.
Available evidence suggests that the immune system of all allograft recipients undergoes clonal expansion.