Fig. 18.1
History of methicillin-resistant Staphylococcus aureus
Looking back at the history of ß-lactam resistance of S. aureus, we notice we have experienced emergence of three generations of MRSA. The first generation appeared just after introduction of methicillin into the community in England in the 1960s [10]. This is now regarded as the Archaic MRSA clone (ST250) [11, 12]. This type spread in Europe and part of the United States, but did not become a pandemic clone. It gradually faded out from Europe by the end of the 1980s and a possible descendant can be found in outbreaks in Spain [13]. In the late 1970s, new outbreaks occurred in the United States and the isolates eventually spread on a global level causing pandemic [14]. We call these isolates second-generation MRSA. In Japan, MRSA isolation started to increase in the 1980s and ST5 is called the NY/Japan clone, which is dominant in healthcare facilities in Japan as well as in the United States. The isolation of MRSA belonging to the two generations, classical and second generation MRSAs, is the only one limited to hospitals and healthcare facilities, and such MRSA is called hospital-acquired or healthcare-associated MRSA (HA-MRSA) [15]. The reason why we started these names is because we started experiencing the emergence of a third generation of MRSA with a different nature [16]. In the 1990s, we started to isolate MRSA from the community and it was called community-acquired MRSA (CA-MRSA) [17–19].
Recently, news that MRSA from pig farms was spreading in communities and causing infection surprised society [20]. A certain clonal group ST398 of MRSA, once the inhabitant of humans, crossed into livestock, where it acquired drug resistance and jumped back to humans. These strains are now called livestock-associated MRSA (LA-MRSA), which is now regarded as another problematic new population in MRSA.
18.2 Emergence of CA-MRSA
In August 1999, the CDC in the United States released a weekly report MMWR with the title of “Four Pediatric Deaths from Community-Acquired Methicillin-Resistant Staphylococcus aureus—Minnesota and North Dakota, 1997–1999” [21]. It reported four cases of fatal MRSA infections. The patients were neonates and infants (12 months to 7 years old). Two cases developed necrotic hemorrhagic pneumoniae and all cases manifested serious septicemia. Those patients did not have any risk factors for MRSA infection. The pattern of drug resistance and pulsed-field gel electrophoresis indicated the isolates were different from those of nosocomial MRSA. By this time, several sporadic cases have been reported in United States. In March 2003, Science posted a short report “Resistant Staph Finds New Niches” describing “MRSA has been an important cause of infections mainly in hospitals and nursing homes, where it’s often resistant to all antibiotics except vancomycin, considered the last resort” [22]. Epidemiologists started seeing more cases of MRSA among people who had no specific connection to hospitals-for instance, among Native Americans, athletic teams, intravenous drug users, and schoolchildren [23–25]. Initially, some researchers speculated that those “community-acquired” microbes might have “escaped” from hospitals. But a genetic comparison of community-and hospital-acquired MRSA strains by Patrick Schlievert of the University of Minnesota, Twin Cities, and colleagues, published in January, suggests that the community variety arose independently and not long ago, when a wild strain picked up a so-called cassette chromosome, a mobile genetic element, containing a gene for methicillin resistance. At that time, several outbreaks of furuncles/carbuncles turned up on hands, legs, chests, buttocks, and genitalia among prisoners and gays had drawn the attention of doctors on the US West Coast [26]. Similar strains were also isolated in Europe and the strong virulence has been focused although the incidence ratio is very low. Most focused elapse of disease starts with skin soft tissue infection and shifting to hemorrhagic necrotic pneumonia [27, 28]. Most isolates produced a leukotoxin called Panton Valentine Leukocidin (PVL) and PVL has been regarded as one of the important virulence factors produced by CA-MRSA [27]. This toxin was discovered from S. aureus causing skin soft tissue infection by Philip Noel Panton and Francis Valentine in 1983 and designated after their names [29, 30]. Not all CA-MRSA produce PVL but PVL is regarded as an important virulence factor of CA-MRSA [31, 32].
In the United States, lineages such as USA300 and USA400 started to cause outbreaks in healthcare facilities suggesting that these CA-MRSA started to invade nosocomial settings and become established [33, 34]. In the near future, classification of MRSA by the site of isolation will be useless. In Japan, there was no sign of increase of CA-MRSA isolation from such severe fatal infection but very recently our laboratory started to receive a number of CA-MRSA from severe SSTI and fatal infections suggesting that the isolation of CA-MRSA is gradually increasing.
18.3 Clinical Background of CA-MRSA Infection and Bacteriology
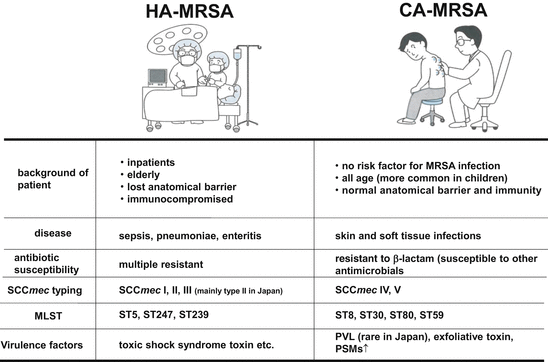
Classification of MRSA into HA-MRSA or CA-MRSA depends on the clinical background of the patient where the isolate derived from and bacteriology of the isolate. Normally CA-MRSA is isolated from healthy personnel unrelated to previous history of medical treatment [28]. More precisely, isolates from the patient such as (1) outpatient or inpatient within 48 h post-admission, (2) no history of isolation of MRSA, (3) no history of operation, renal transfusion, hospitalization, (4) no history of indwelling of catheter or medical devices are regarded as CA-MRSA. Also CA-MRSA possesses several bacteriological characteristics as shown in Fig. 18.2. In Japan, HA-MRSA carries SCCmec type II or III and often manifests multidrug resistance: HA-MRSA is resistant to not only ß-lactams but macrolides and aminoglycosides [35]. On the other hand, CA-MRSA carries SCCmec type IV or V, which is a relatively smaller sized element and susceptible to other antimicrobial agents [36]. But USA300 is an exception: it shows resistance to fluoroquinolones and mupirocin as well [37]. USA300 is a multidrug resistant CA-MRSA now prevalent worldwide and in Japan the isolation started after 2007 [38].
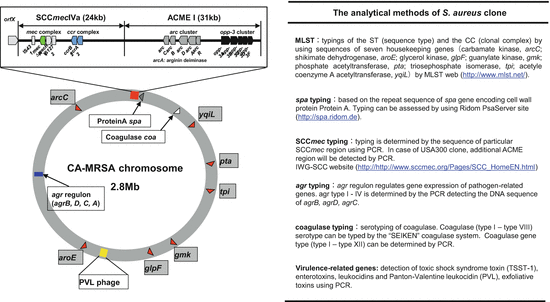

Fig. 18.2
Genotyping methods of MRSA. Left, schematic genome map of CA-MRSA. Grey squares indicate housekeeping genes. Right, the genetic typing methods to analyze S. aureus clone
18.4 Genetic Diversity of CA-MRSA
Upon acquiring SCCmec, S. aureus becomes MRSA [39]. SCCmec is a mobile genetic element flanked by att sites of about 15 bp nucleotide sequences in both ends. SCCmec is integrated into the genome through recombination at the att sequence present in the 3′ terminal of orfX [40, 41]. SCCmec can be classified into 11 groups, I–XI [41, 42]. As shown in Fig. 18.3, SCCmec is composed of the ccr gene complex (ccr: cassette chromosome recombinase involved in site-specific recombination of SCCmec) and mec gene complex. Typing is stipulated by the International Working group on the Staphylococcal Cassette Chromosome elements: IWG-SCC (http://www.sccmec.org) [43, 44]. The ccr gene complex is composed of either ccrAccrB or ccrC alone. ccrAccrB is classified into four different types (AB1, AB2, AB3, AB4) according to sequence difference [45]. The mec gene complex is composed of mecA and upstream two regulatory genes, mecR1 and mecI, and is classified into three types, classes A–C [11, 46]. In class A, mecR1 and mecI are present just upstream of mecA and IS431 downstream of mecA. In class B, mecR1 is truncated by insertion of IS1273. In class C, mecR1 is truncated by IS431 in the same (C1) or opposite (C2) direction to the preceding IS431 forming a transposon.
Emergence of MRSA started with Archaic clone (ST250/SCCmec I) [47, 48]. Then the second generation pandemic clones including the New York/Japan clone (ST5/SCCmec II) [35], pediatric clone (ST5/SCCmec IVa) [49], Berlin clone (ST45/SCCmec I) [15, 50, 51], Iberian clone (ST247/SCCmec I) [48, 51], Brazilian clone (ST239/SCCmec IIIa) [52, 53], EMRSA-15 (ST22/SCCmec IV) [54, 55], EMRSA-16 (ST36/SCCmec II) [49, 56], and others (Fig. 18.4). CA-MRSA appeared worldwide around 1999 as PVL-producing virulent MRSA. Representative PVL-positive CA-MRSA include USA400 (ST1/SCCmec IVa) [57] and USA300 (ST8/SCCmec IVa) prevalent in the United States [37], European clone (ST80/SCCmec IV) [58], pandemic clone (ST30/SCCmec IVa or IVc) [59], the clone prevalent in Taiwan and United States (ST59/SCCmec V) [60, 61], new European clone (ST22/SCCmec IV or V, in Japan IVa) [62, 63], and Bengal Bay clone (ST772/SCCmec V) prevalent in India and Malaysia [64, 65]. In Japan, the ratio of isolation of PVL-positive CA-MRSA is extremely low, 3–5 % compared to that of the United States [66]. CA-MRSA in Japan is largely PVL-negative. Recently, ST8/SCCmec IV distinct from the USA300 clone has been frequently isolated from pneumoniae, sepsis, musculoskeletal abscess, and disseminated severe infection [67, 68]. This clone carries spj encoding new cell wall protein CWASP/J in SCCmec in addition to tst and sec although this does not possess pvl and ACME genes (Fig. 18.5) [67, 69]. More classical CA-MRSA belonging to ST88, ST89, and ST91 produces exfoliative toxin B and is involved in impetigo and staphylococcal scalded skin syndrome in neonate/infants [35]. Genetical relations of representative clonal complexes of HA-MRSA, CA-MRSA, and recently emerged LA-MRSA are depicted in Fig. 18.6. Table 18.1 summarizes the genetic diversity of the CA-MRSA clone in the world.
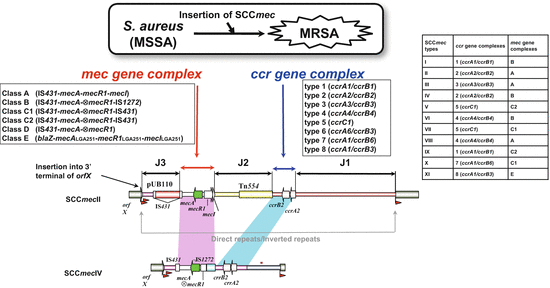
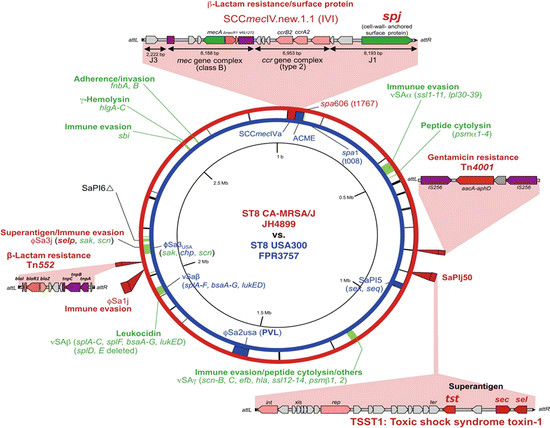
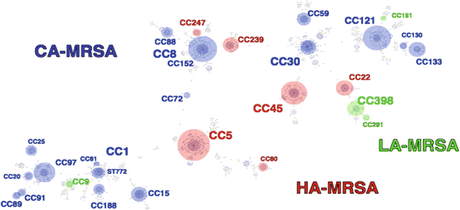

Fig. 18.4
Structure and typing of SCCmec. Methicillin-susceptible S. aureus will be converted to MRSA upon acquiring SCCmec, a genomic island encoding mecA and other genetic elements. SCCmec carries several elements including (1) the mec gene complex (mec); (2) the ccr gene complex (ccr); (3) regions other than mec and ccr designated as “joining regions” (J1, J2, and J3); (4) characteristic nucleotide sequences, inverted repeats, and direct repeats at both ends; and (5) the integration site sequence (ISS) for SCC. The SCCmec is located at the 3′ end of orfX in chromosome

Fig. 18.5
Genomic comparison of ST8/SCCmecIVl and USA300. Genomic information of JH4899 (ST8/SCCmecIVl) and USA300 (ST8/SCCmecIVa) are presented in circular maps (outside, JH4899; inside, USA300). Note a novel cell wall protein CWASP/J encoded by the spj in SCCmecIVl. JH4899 lacked phiSA2usa (carrying a PVL gene) and ACME, which are present in USA300. (Part of figure was modified from Ref. [68])

Fig. 18.6
Clonal diversity of MRSA by e-BURST analysis. eBURST analysis using MLST indicated relations of HA-MRSA, red; CA-MRSA, blue; and LA-MRSA, green
Table 18.1
Diversity of CA-MRSA clones
ST | arcC | aroE | glpF | gmk | pta | tpi | yqiL | spa | SCCmec | Regional clones |
|---|---|---|---|---|---|---|---|---|---|---|
1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 131 | IV | USA300, CMRSA7, WA-1 |
5 | 1 | 4 | 1 | 4 | 12 | 1 | 10 | 2 | I, II, IV, V, VI | Geradine, Pediatric |
6 | 12 | 4 | 1 | 4 | 12 | 1 | 3 | Unknown | IV | – |
8 | 3 | 3 | 1 | 1 | 4 | 4 | 3 | 1 | IV, V, VI | USA300, CMRSA10, WA-12 |
22 | 7 | 6 | 1 | 5 | 8 | 8 | 6 | 113 | IV, V | – |
30 | 2 | 2 | 2 | 2 | 6 | 3 | 2 | 19 | IV, V | SWP, OSPC, WSPP, USA1100 |
45 | 10 | 14 | 8 | 6 | 10 | 3 | 2 | 73 | IV, V, VI | – |
59 | 19 | 23 | 15 | 2 | 19 | 20 | 15 | 17 | IV, V | USA1000, Taiwan |
72 | 1 | 4 | 1 | 8 | 4 | 4 | 3 | 193 | IV, V | USA700 |
75 | 36 | 3 | 43 | 34 | 39 | 52 | 49 | Unknown | IV, V | WA-8, WA-79 |
78 | 22 | 1 | 14 | 23 | 12 | 53 | 31 | Unknown | IV | – |
80 | 1 | 3 | 1 | 14 | 11 | 51 | 10 | 70 | IV | European CA-MRSA |
88 | 22 | 1 | 14 | 23 | 12 | 4 | 31 | 9 | IV, V, VI | – |
91 | 1 | 26 | 28 | 18 | 18 | 54 | 50 | 416 | II, IV, V | – |
93 | 6 | 64 | 44 | 2 | 43 | 55 | 51 | 1143 | IV, V | Queensland |
97 | 3 | 1 | 1 | 1 | 1 | 5 | 3 | 105 | IV, V | – |
121 | 6 | 5 | 6 | 2 | 7 | 14 | 5 | 312 | V | – |
152 | 46 | 75 | 49 | 44 | 13 | 68 | 60 | 207 | V | Balkan |
377 | 46 | 75 | 49 | 50 | 13 | 68 | 60 | 207 | V | Balkan |
772 | 1 | 1 | 1 | 1 | 22 | 1 | 1 | 692 | V | Bengal Bay, WA-60 |
18.5 Virulence Factors of CA-MRSA
When bacteria attach and adhere to the surface of a host, and invade into the body, the host will recognize the bacteria as a foreign body and start exerting an eliminating system, which is called immunity. S. aureus produces a variety of virulence factors including cell surface proteins and secreted protein toxins, and the degree of production and their repertoire vary according to clone types (Fig. 18.7). The bacteria also possess evasion tools to escape from the host immune system [70, 71]. HA-MRSA is associated with inpatients in hospitals/health care facilities [72]. Therefore it might be relatively easy for them to go inside the host body inasmuch as all they have to do is to use the anatomical barrier niche such as catheter tubing to invade the blood circulation and start disrupting the acquired immunity system using superantigens. On the other hand, CA-MRSA infects healthy bodies and it needs to go through a tight anatomical barrier to reach soft tissue. Therefore CA-MRSA needs a variety of virulence factors necessary to overcome the innate immune barrier system. Some important virulence factors for CA-MRSA are listed as follows.
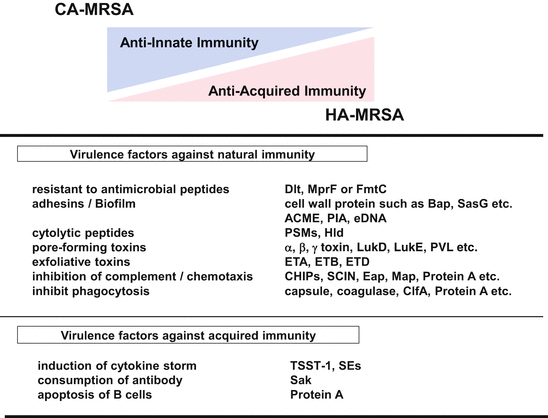

Fig. 18.7
Comparative characterization of virulence factors of HA-MRSA and CA-MRSA
18.5.1 Leukocidin
Several types of leukocidins are known including PVL that selectively lyse leukocytes and its variants LukED and LukMFPV, and Luk that lyses rabbit red blood cells [73, 74]. PVL production is reportedly associated with furunculosis [75]. Genes for PVL are tandemly encoded as lukS-PV and lukF-PV forming an operon and the genes are embedded in the lysogenic phage genome between the lytic enzyme gene and att site of the phage [74]. This phage can mobilize genes for PVL and several PVL-transducing phages have been reported such as ФPVL and ФSTL [76, 77]. PVL is composed of two components, LukS-PV and LukF-PV, and these components attack and integrate into the cell membrane of white blood cells (neutrophils, granulocytes, monocytes, macrophages, etc.) to form pores made of hexamer–octamer, which induce necrosis in the end [74, 78, 79]. Under low concentration, PVL induces apoptosis or the generation of inflammatory factors such as IL-8 and LTB4 [80, 81]. PVL is suggested to be involved in the onset of skin and soft tissue infection such as furunculosis and carbuncle [82]. PVL-producing S. aureus often causes invasive infectious diseases such as deep-seated subcutaneous abscess, osteomyelitis, necrotic pneumoniae, and sepsis [25]. PVL is one of the leading virulence factors involved in CA-MRSA infection, but some cases by PVL-nonproducing CA-MRSA exist. In the case of severe invasive CA-MRSA infection, a PVL-producing strain must be taken into account for the etiological agent.
18.5.2 Cytolytic Peptides
Cytolytic peptides are a complex of peptides that are produced by virtually all MRSA isolates. They are called PSMs (phenol soluble modulin-like peptide) based on their solubility to phenol [83]. Each peptide is N-terminally formulated. Various groups are known including δ hemolysin composed of 26 amino acids, four types of PSMαs, PSMα1-4 composed of 20–25 amino acids, and two types of PSMβs, PSMβ1-2 composed of 44 amino acids. CA-MRSA is reported to produce more abundant PSMs than HA-MRSA [25, 84]. PSMs are suggested to induce lysis of neutrophils, production and disruption of biofilms, and are implicated in pathogenesis of CA-MRSA infection [83]. In HA-MRSA, a gene for PSM-mec, 20 amino acids peptide, is encoded in the J2 region of SCCmec II, III, and VIII [85]. A recent study indicated a transcript of PSM-mec acts as a functional psm-mec RNA suppressing other pathogenic genes on the genome and renders HA-MRSA less virulent [86, 87]. In support of this, a point mutation was found in the promoter region of psm-mec and this mutation canceled the suppression of genes for PSMαs and induced high expression of PSMαs in highly virulent HA-MRSA (SCCmec II).
18.5.3 Arginine Catabolic Mobile Element (ACME)
ACME was first discovered in CA-MRSA USA300 (ST8/SCCmecVa) as the complex of SCCmecVa (ca. 34 kb) and ACME (ca 31 kb), but later found in ST5/SCCmec II and IV, ST59/SCCmecIVa, ST97/SCCmec V, ST1/SCCmec Iva, and ST239/SCCmec III, and coagulase negative staphylococci such as Staphylococcus epidermidis, S. haemolyticus, and S. capitis [37, 88–90]. ACME is a mobile element of a gene cluster composed of two clusters, the arginine–metabolism gene (arc) cluster (arcA, arcB, arcD, arcA, arcR) and the quorum-sensing gene cluster (opp-3A, opp-3B, opp-3C, opp-3D, opp-3E) [37]. There are three types I–III in ACME: ACME-I possesses clusters of arc and opp; ACME II possesses only the arc cluster; ACME-III possesses only the opp cluster [37, 91, 92]. Involvement of ACME to adherence to skin and mucosal surface, and horizontal transfer of the bacteria have been suggested but detailed mechanisms remain elusive. The arc cluster is involved in the metabolic pathway of arginine (arginine deiminase system, ADS) and ornithine, ammonia, carbon dioxide, and ATP are generated as the metabolites of L-arginine [93]. The generation of ammonia by ADS neutralizes the acidic condition of skin thereby enabling colonization of S. aureus on the skin surface. The Opp-3 cluster is implicated in the uptake of peptides as nutrient and in regulation of growth through quorum-sensing [94].
18.5.4 Attachment and Adherence to Host (Biofilm)
S. aureus uses two mechanisms for their adherence to host: nonspecific binding by using hydrophobic and/or electrostatic interaction with plastic or metal surface; and specific binding to extracellular matrix and/or serum protein [95]. Bacterial proteins used for such binding are collectively called microbial surface components recognizing adhesive matrix molecules (MSCRAMM). MSCRAMM includes proteins such as Protein A (Spa), fibronectin-binding protein (FnBPA/B), collagen-binding protein (Cna), fibrinogen-binding protein (ClfA/B), Sdr protein, and others, and they are believed to be involved in initial attachment of the bacteria to the tissue [96]. Subsequent adhesion and biofilm formation are necessary for the bacteria to establish infection in the host [97]. Polysaccharide intercellular adhesion (PIA) is the major such factor, and genes involved in synthesis and production of PIA are encoded on icaA, icaD, icaB, and icaC as an operon [98–100].
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree