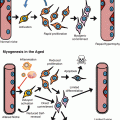
Fig. 2.1
Schematic for yeast replicative lifespan (RLS) and chronological lifespan (CLS). Replicative lifespan is defined by the number of daughter cells a mother cell can produce prior to senescence. Chronological lifespan is defined by the length of time that a population of nondividing yeast cells can maintain replicative potential when restored to rich media
2.2 Replicative Lifespan
The budding yeast Saccharomyces cerevisiae divides asymmetrically with the original “mother” cell budding to give rise to a “daughter” cell (Barker and Walmsley 1999; Jazwinski 1990). The first replicative lifespan studies (RLS) were published more than 60 years ago by Mortimer and Johnston, who designed a method to measure the number of buds or daughters produced by a single mother cell (Mortimer and Johnston 1959). Based on continual observation of individual cells and separation of the morphologically asymmetric buds, they determined that the number of buds produced by one “mother” cell is limited, defined as its lifespan. After undergoing a certain number of mitotic divisions, cells cease to divide and enter a short post-replicative state followed eventually by cell lysis.
The RLS assay is traditionally performed using a standard dissection microscope with a micromanipulator. From a logarithmically growing culture, individual newborn daughter cells are isolated onto a solid-media substrate. Subsequent budded daughter cells are separated manually from mother cells by microdissection, and the number of divisions each mother cell has produced is then recorded, the totals of which result in the mean RLS (Kaeberlein and Kennedy 2005; Steffen et al. 2009). The typical mean lifespan of laboratory wild-type Saccharomyces cerevisiae is around 25 generations, with the maximum being around 40 (Gillespie et al. 2004). It is important to note that strain background, nutritional conditions, and temperature can have major effects on the outcome of RLS experiments (Kirchman et al. 1999; Kaeberlein and Powers 2007; Smeal et al. 1996; Kaeberlein 2010; Shama et al. 1998). While this technique may be the most accurate for determining lifespan, it has limitations including being labor intensive and suffers from an inability to generate large populations of old cells for biochemical analysis.
Lindstrom and Gottschling developed a new method, the Mother Enrichment Program, which blocks genetically the proliferative potential of daughter cells (Lindstrom and Gottschling 2009). This method can provide a large number of aged mother cell cohorts at a late stage of their lifespan, allowing the subsequent analysis of age-associated phenotypes, such as gene expression or protein levels. Recently, microfluidic devices have been developed to study yeast aging, providing another high-throughput method for automated analyses of single cells. These microfluidic systems use tiny transparent chambers to trap single mother cells while automatically removing daughter cells by continuous medium flowing through the device and allow for high-resolution microscopic live-cell imaging system to capture a cell throughout the aging process (Lee et al. 2012; Xie et al. 2012; Zhang et al. 2012). Together, these techniques now provide the capacity for a wide range of experimental approaches to study the yeast replicative aging process.
An early hypothesis to explain the limited lifespan of yeast cells was based on the observation that permanent, non-overlapping bud scars remained on the cell surface after each division (Barton 1950). In addition, cells increase in size as they get older, which reduces surface-to-volume ratios. Thus, a reduced relative surface area may eventually impair division of aging cells (Bacon et al. 1966; Seichertova et al. 1975). A more recent version, the “hypertrophy” hypothesis, has been proposed whereby yeast cells stop dividing after reaching a maximum size (Yang et al. 2011; Bilinski 2012; Bilinski et al. 2012). Longer-lived mutants would start out smaller as buds or increase in size at a reduced rate during the budding process. However, this theory has been disputed and remains controversial (Ganley et al. 2012; Kaeberlein 2012).
Since the early 1990s, when it was demonstrated that genetic manipulation were sufficient to alter yeast replicative lifespan (D’Mello et al. 1994; Kennedy et al. 1995; Sun et al. 1994), the yeast aging field has become a prominent model for aging research, serving as a platform for the discovery and characterization of genes, pathways, and molecular mechanisms involved in aging. As a result, there is growing interest in understanding which characteristics of replicative aging in yeast are shared with mammals, either with respect to the lifespan of the organism or the survival and proliferative capacity of adult cells within the mammalian organism.
2.3 Dietary Restriction and Yeast Replicative Lifespan
Dietary restriction (also referred to as calorie restriction) is defined as a reduction in nutrient intake without malnutrition. Even though the first demonstration of lifespan extension by dietary restriction (DR) was 80 years ago in rats (McCay et al. 1989), the mechanism behind its effect is still only partially understood (Gallinetti et al. 2013; Gems and Partridge 2013; Guarente 2013). DR is the only environmental intervention that can extend lifespan in all common model organisms, including yeast, worm, flies, mice, primates, and several others (Fontana et al. 2010; Anderson and Weindruch 2010; Masoro 2005; Weindruch et al. 1986; Colman et al. 2009; Mattison et al. 2012). In yeast, DR is typically invoked by reducing levels of glucose in the media from 2 % to either 0.5 % or 0.05 % during the lifespan experiment (Lin et al. 2000; Kaeberlein et al. 2004a). DR has been shown to increase both replicative and chronological lifespan in yeast (Kaeberlein 2010; Kennedy et al. 2007); however, the mechanism(s) proposed to explain the effects of DR in yeast remains only partly understood (Steinkraus et al. 2008). HXK2 encodes the glycolytic enzyme hexokinase II, which converts glucose to glucose-6-phosphate and leads to glucose entering the glycolytic pathway (Walsh et al. 1983). Genetic models of DR, such as deletion of HXK2 and other nutrient-responsive genes, also lead to RLS extension (Lin et al. 2000). Amino acid restriction, while maintaining normal glucose levels, has also been shown to extend lifespan (Jiang et al. 2000). However, the contribution of each amino acid component to aging is still a mystery. Methionine restriction has been shown to improve lifespan from yeast to rodents (Orentreich et al. 1993; Richie et al. 1994; Johnson and Johnson, 2014). A recent study showed that specific amino acid and glucose affect aging through different pathways (Mirisola et al. 2014). They demonstrated that threonine and valine promoted aging and sensitized yeast cells to stress mainly by activating the Tor/S6K pathway, while serine promoted aging by activating sphingolipid-mediated serine protein kinase (Pkh1/2); glucose promoted aging and sensitized yeast cells to stress through a Ras-dependent mechanism (Mirisola et al. 2014). Moreover, the function of amino acid transporter showed to link with longevity in yeast. A recent study reported that the inhibition of tryptophan uptake and destabilization of tryptophan permease Tat2 are benefit for increasing yeast RLS (He et al. 2014).
Much of the focus on the mechanisms by which DR extends lifespan has centered on the silent information regulator 2 (SIR2), which belongs to the deacetylase sirtuin protein family. SIR2 and its role in RLS are discussed in greater detail later in this chapter. It was originally proposed that DR increased RLS by activation of Sir2p (Lin et al. 2000); however, this hypothesis has since been called into question as DR was found to extend lifespan via Sir2-independent mechanisms (Kaeberlein et al. 2004b). Whether other Sirtuins play a redundant role with Sir2p in DR-mediated lifespan extension has also been debated (Lamming et al. 2005; Tsuchiya et al. 2006). Understanding the role of SIR2 in yeast aging is important given the findings implicating SIRT1, the mammalian ortholog, in aging, stem cell function, and cellular senescence (Giblin et al. 2014; Imai and Guarente 2014).
More recently, there has been a growing recognition of the possibility that lifespan extension by DR is mediated via several partially redundant nutrient-responsive signal transduction pathways, including PKA, TOR, and SCH9. Mutations with reduced PKA, TOR, or SCH9 signaling are replicatively long lived, and lifespans of these mutants cannot be further extended by DR (Kaeberlein et al. 2005c; Lin et al. 2000; Fabrizio et al. 2004). While the TOR pathway has been linked to Sir2 function (Ha and Huh 2011), the totality of evidence suggests that reduced signaling through these nutrient-responsive pathways occurs through multiple mechanisms (Longo et al. 2012).
Yeast cAMP-dependent protein kinase (PKA) is a conserved serine/threonine kinase complex containing three catalytic subunits encoded by TPK1, TPK2, and TPK3 (Toda et al. 1987a, b). In yeast, cAMP/PKA is a major signaling pathway for growth on different carbon sources. The basal level of cAMP is higher in yeast growing on fermentable carbon sources (glucose, fructose, and sucrose), compared to growing on non-fermentable carbon sources (glycerol, ethanol, and acetate) (Robertson et al. 2000). PKA is regulated by at least two upstream sensing pathways via the induction of cAMP (Santangelo 2006). One sensing pathway involves RAS activation (Busti et al. 2010). G-protein Ras stimulates acetylate cyclase (Cyr1), which leads to increased cAMP levels in yeast (Thevelein 1994). RAS activation is upregulated by guanine nucleotide exchange factors such as Cdc25p (Martegani et al. 1986). Prevention of activation of this pathway by deletion of CDC25 leads to lifespan extension (Lin et al. 2000). The other upstream sensing pathway is a G-protein-coupled receptor system encoded by GPA2 and GPR1. Deletion of either GPA2 or GPR1 leads to RLS extension, and these mutants are commonly used as models for reducing PKA activity in lifespan experiments (Lin et al. 2000).
Another major nutrient-responsive pathway implicated in yeast replicative aging is the target of rapamycin (TOR) network (Heitman et al. 1991). The TOR protein is a serine/threonine kinase conserved in all eukaryotes (De Virgilio and Loewith 2006). Unlike higher eukaryotes, yeast has two TOR proteins, Tor1p and Tor2p, which function in two distinct multiprotein complexes: TOR complex 1 (TORC1) and TOR complex 2 (TORC2) (Dann and Thomas 2006; Loewith et al. 2002). TORC1 contains either Tor1p or Tor2p and is sensitive to rapamycin (Loewith et al. 2002; Stan et al. 1994). TORC2 contains only Tor2 (Loewith et al. 2002). TORC2 regulates polarization of the actin cytoskeleton in a rapamycin-insensitive manner (Zheng et al. 1995; Loewith et al. 2002). Aside from rapamycin, evidence demonstrates that the TOR signaling pathway is regulated by the presence of nitrogen sources such as glutamine availability; however, the sensing mechanism is still unclear (Crespo et al. 2002; Carvalho and Zheng 2003).
TOR was proposed as a primary conduit in RLS regulation based on the analysis of replicative lifespan in 564 single-gene deletion strains of yeast (Kaeberlein et al. 2005c). Among the ten single-gene deletions with increased RLS, which were identified from the 564 randomly selected deletion strains, at least five of these correspond to gene-encoding components of the TOR pathway (Kaeberlein et al. 2005c). Reduced TOR signaling by either rapamycin treatment or in a tor1Δ mutant strain leads to RLS extension in yeast (Kaeberlein et al. 2005c; Medvedik et al. 2007a). DR treatment to the tor1Δ mutant fails to further increase lifespan, indicating that TOR regulation of lifespan is through an overlapping pathway with DR (Kaeberlein et al. 2005c). Reduced TOR activity also leads to lifespan extension in C. elegans (Vellai et al. 2003; Jia et al. 2004; Hansen et al. 2008), D. melanogaster (Kapahi et al. 2004; Bjedov et al. 2010), and mice (Komarova et al. 2012; Harrison et al. 2009; Anisimov et al. 2011), suggesting that TOR signaling not only plays an important role in mediating the longevity effects of DR but also is a conserved nutrient-responsive pathway throughout divergent eukaryotic species.
Sch9p is also a nutrient-responsive kinase that can be phosphorylated by Tor1p and is the functional ortholog of mammalian ribosomal S6 kinase (Powers 2007; Urban et al. 2007). SCH9 is required for TORC1-mediated regulation of ribosome biogenesis, translation initiation, and entry into G0 phase (di Blasi et al. 1993; Hay and Sonenberg 2004). Sch9p also shows sequence homology to the kinase Akt, a central component of insulin/IGF-1-like signaling pathways (Paradis and Ruvkun 1998; Burgering and Coffer 1995). Even though yeast lacks a formal insulin/IGF-1-like signaling pathway, Sch9 may have a comparable role to both Akt and S6 kinases (Steinkraus et al. 2008). Deletion of SCH9 extends both mean and maximum RLS (Fabrizio et al. 2004; Kaeberlein et al. 2005c) in yeast, and consistent with this observation, deletion of either Akt homologs or S6 kinase extends lifespan in C. elegans (Pan et al. 2007; Hansen et al. 2007; Paradis and Ruvkun 1998). Mice lacking S6 kinase 1 (S6K1) also have enhanced longevity (Selman et al. 2009). Interestingly, a recent report suggested that independently of TORC1, Sch9p can be independently phosphorylated and activated by the Snf1 kinase, which is the yeast ortholog of the mammalian AMPK kinase, through the acetylation of the Snf1 complex component, Sip2p (Lu et al. 2011). Enhanced Sip2p acetylation extends lifespan, likely because it results in downregulation of Snf1p-mediated phosphorylation of Sch9p (Lu et al. 2011).
Increasing attention has been paid to the downstream mediators of the PKA, TOR, and SCH9 pathways, specifically with regard to their importance in yeast RLS. Recently, a comprehensive, computational network of TORC1 has been constructed (Mohammadi et al. 2013). The functional map of TOR downstream effectors can be used to predict transcriptional changes and posttranslational modifications in response to TOR inhibition, which helps to identify new targets for antiaging therapy (Mohammadi et al. 2013).
Regulation of mRNA translation has been shown to be one of the most relevant downstream mediators required for RLS extension. It has been repeatedly shown that decreased mRNA translation, either by downregulating ribosomal protein biosynthesis or translation initiation factors, leads to lifespan extension (Steffen et al. 2008). The yeast ribosome has a small (40S) and a large (60S) ribosomal subunit which contains 78 ribosomal proteins (RP) encoded by 137 RP genes (Kaeberlein 2010). Although most RP subunits are essential in yeast, the majority of RP genes are duplicated, which allows for a viable deletion of one of the paralogs (Komili et al. 2007; McIntosh and Warner 2007). In the initial yeast deletion collection screen for replicative lifespan, the rpl31aΔ and rpl6bΔ strains, which lack genes coding for ribosomal large subunit proteins, were identified as long lived, although not every RP deletion extends lifespan (Kaeberlein et al. 2005c). Follow-up work demonstrated that among the 20 RP gene deletions which have been shown to increase RLS, 18 genes encode the 60S-subunit proteins, indicating genes that encode the components of the 60S subunit appear to more robustly regulate yeast RLS (Chiocchetti et al. 2007; Kaeberlein et al. 2005c; Managbanag et al. 2008; Steffen et al. 2008, 2012). Deletion of any one of three mRNA translation initiation factors (tif1Δ, tif2Δ, and tif4631Δ) has also been found to confer RLS extension (Steffen et al. 2008). Again, lifespan regulation by ribosomal proteins can also be found in another eukaryotic species. In C. elegans, an RNAi screen demonstrated that decreased mRNA translation level is associated with enhanced lifespan (Pan et al. 2007; Chen and Contreras 2007; Curran and Ruvkun 2007; Hansen et al. 2007), suggesting that modulation of mRNA translation by reducing the TOR signaling pathway is a conserved mechanism throughout divergent eukaryotic species (Smith et al. 2008).
Gcn4p is another downstream mediator of the nutrition signaling pathway. Gcn4p is one of the main transcriptional activators of amino acid biosynthetic genes, specifically in response to amino acid starvation (Hinnebusch 2005; Hinnebusch and Natarajan 2002). Gcn4p abundance is translationally regulated by four small inhibitory upstream open reading frames (uORFs) in the 5′ untranslated region of the GCN4 mRNA (Hinnebusch 2005). Due to the uORFs, GCN4 is largely not translated under normal growth conditions. Nevertheless, alterations in 60S ribosomal subunit levels lead to increased translation of GCN4, despite an overall decrease in protein synthesis. This evidence suggests that decreased mRNA translation affects aging at least in part by differentially altering the translation of specific mRNA targets. As a result of these findings, Gcn4 has been identified as a translational regulator responsible for the role of the 60S subunit with regard to lifespan regulation, although GCN4-independent effects are also apparent (Steffen et al. 2008). In addition, yeast RLS extension resulting from DR, tor1Δ, and sch9Δ can be partially blocked by gcn4Δ, indicating that GCN4 is a downstream mediator of DR, TOR, and SCH9 in a conserved aging pathway. This pathway might also be conserved in mammals as recent evidence indicates that protein levels of the mammalian ortholog of GCN4, ATF4, are increased in fibroblasts from liver tissue from long-lived mouse models (Li and Miller 2015).
2.4 Sirtuins
Sirtuins are a highly conserved family of nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases. Sirtuins have been well studied in recent years for their potential roles in the regulation of yeast lifespan and because of their highly conserved nature (Finkel et al. 2009; Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). Sirtuins are involved in multiple physiological processes including cellular stress resistance, genomic stability, energy metabolism, and longevity (Wierman and Smith 2014). Moreover, reports have shown that sirtuin activators are potentially beneficial in the treatment of mammalian disease models (Finkel et al. 2009; Michan and Sinclair 2007; Guarente 2006; Imai and Guarente 2010).
The silent information regulator (SIR) genes, including SIR2, SIR3, and SIR4, were first identified in S. cerevisiae as being required for silencing at the cryptic mating-type loci, HML and HMR (Ivy et al. 1986; Rine and Herskowitz 1987). In S. cerevisiae, Sir2p is a vital component of multiple protein complexes that are required for silencing chromatin regions in telomeres and ribosomal DNA (rDNA; Aparicio et al. 1991; Guarente 1999; Smith and Boeke 1997). It has been well established that Sir2p is recruited to the telomeres by Sir3p and Sir4p, as well as other factors. Disruption of these proteins leads to loss of silencing of the telomeres (Koch and Pillus 2009). Also, Sir2p protein abundance declines with age (Dang et al. 2009), which is consistent with findings that an extra genomic copy of SIR2 leads to lifespan extension (Kaeberlein et al. 1999). Moreover, the decrease in Sir2 protein abundance correlates with an increase in histone H4 lysine 16 acetylation and loss of histones near telomeres. When histones are lost from the telomeres, transcriptional silencing near the telomeres is also lost. Overexpression of histones extends lifespan (Dang et al. 2009; Feser et al. 2010). Together, this data suggests that the Sir2p-dependent stabilization of telomere chromatin could have a critical role in RLS. Due to the potential relationship between telomeric dysfunction, cell senescence, and aging in humans, the hypothesis relating Sir2p to telomere function in yeast is appealing (Shawi and Autexier 2008; Blackburn et al. 2006).
S. cerevisiae contains 100–200 copies of an rDNA repeat unit, which are located in tandem on chromosome XII (Johnston et al. 1997; Petes 1979). The rDNA tandem array is unstable, with repeat excision from the genome occurring by homologous recombination and leading to formation of extrachromosomal rDNA circles (ERCs) (Sinclair and Guarente 1997). ERCs replicate during S-phase, but are largely maintained in mother cells during cell division because they do not have a centromeric element. As a result, they accumulate to toxic levels in the mother cell with age and can limit their lifespan (Sinclair and Guarente 1997). It has been hypothesized that Sir2p activity promotes replicative aging by suppressing ERC formation (Sinclair and Guarente 1997). Mutants lacking SIR2 display a six- to tenfold increase in rDNA recombination and have a 50 % reduced lifespan. On the other hand, overexpressing SIR2, through integration of a second SIR2 copy, extends replicative lifespan (Kaeberlein et al. 1999, 2004a; Kennedy et al. 1995). Additionally, Fob1 promotes ERC formation as an indirect consequence of its role in regulating replication fork traffic in the rDNA (Kobayashi and Horiuchi 1996). Mutants lacking FOB1 display a dramatic reduction in rDNA recombination and ERC levels and a significant increase in replicative lifespan (Defossez et al. 1999). fob1Δ rescues the short replicative lifespan seen in sir2Δ (Kaeberlein et al. 1999; Defossez et al. 1999). Both sir2Δ fob1Δ and fob1Δ strains have low ERC levels, suggesting that FOB1 is epistatic to SIR2 for rDNA recombination (Kaeberlein et al. 1999). However, the lifespan of the fob1Δsir2Δ strain is similar to wild type, and shorter than that of the fob1Δ strain, indicating that Sir2 has a second function for regulating lifespan which is independent of both Fob1 and ERC accumulation (Kaeberlein et al. 1999). A recent study also showed that age-associated ERC formation can be FOB1 independent using Mother Enrichment Program (Lindstrom et al. 2011). This study reported that increased rate of homologous recombination of rDNA, rather than ERCs, contributes to aging in yeast (Lindstrom et al. 2011). It is also suggested that Sir2 protein levels are not the major factor regulating rDNA recombination in old cells.
Inhibition of TOR signaling by DR or by rapamycin treatment increases rDNA stability by enhancing the association of Sir2p with rDNA (Ha and Huh 2011). Recently, a quantitative trait locus (QTL) study shows that SIR2 and rDNA loci play predominant roles in RLS (Stumpferl et al. 2012). Another QTL study also indicates that polymorphisms of rDNA, independent of SIR2 and FOB1, contribute to the lifespan extension due to modulating genome replication dynamics (Kwan et al. 2013). It has also been reported that Sir2 modulates lifespan through repression of E-pro, an rDNA noncoding promoter (Saka et al. 2013). When E-pro is repressed by an inducible promoter, it shows increased rDNA stability, decreased ERC levels, and increased RLS. Moreover, Sir2 is dispensable for lifespan extension when E-pro is repressed, indicating that Sir2 regulates lifespan through the repression of E-pro noncoding transcription in the rDNA loci. Although ERCs have been reported in higher eukaryotes including human tissues, their role in aging process is unknown (Cohen et al. 2010; Cohen and Segal 2009). Another possibility in mammals is that SIRT1 might regulate recombination in other repeated regions of the human genome, which could impact both cell senescence and aging (Oberdoerffer et al. 2008).
There is also evidence that SIR2 has functions outside of modulating rDNA recombination and telomere dynamics. First, it has been reported that Sir2 prevents daughter cells from inheriting oxidative damage that accumulates in aging mothers (Erjavec and Nystrom 2007; Aguilaniu et al. 2003). This hypothesis is further supported by the finding that SIR2 overexpression rescues the short lifespan of yeast treated with H2O2 (Oberdoerffer et al. 2008). Overexpression of Hsp104, a stress tolerance factor that functions with chaperones to dissipate protein aggregation, extends the lifespan of sir2Δ strains (Erjavec et al. 2007). Increased Hsp104 activity also rescues the missegregation of damaged proteins to daughters in the sir2Δ strain. In 2010, Liu et al. showed that Hsp104-recruiting protein aggregates move back from the bud to the mother cell along actin cables in an Sir2-dependent manner (Liu et al. 2010). Because oxidative damage has been implicated in aging process in invertebrates and mammals, there has been a growing interest in studying the link between sirtuins and oxidative stress and their roles in mediating lifespan. In addition, it has been showed that SIRT1, one of the sirtuin members that have been identified in mammals, regulates an oxidative stress response by deacetylating several transcription factors that regulate antioxidant genes (Brunet et al. 2004; Motta et al. 2004; van der Horst et al. 2004). Identifying yet another potential function of Sir2 relevant to aging, a recent study showed that Sir2 exhibits co-enrichment to various chromatin targets involved in metabolism and protein translation (Li et al. 2013).
It has been suggested that DR mediates lifespan in a sirtuin-dependent manner in yeast, worms, flies, and mice, although the topic is still highly debated (Guarente 2005; Rizki et al. 2011; Viswanathan and Guarente 2011; Burnett et al. 2011). One complicating factor is that DR can be induced in different ways and might require different downstream factors in different protocols, as appears to be the case in worms.
In mammals, there are seven sirtuins (SIRT1-7) that function as NAD-dependent deacetylases, deacylases, and/or ADP-ribosyl-transferases (Houtkooper et al. 2012). In mice, SIRT1 is the closest ortholog to yeast SIR2, and it has been suggested to be required for lifespan extension by DR (Dryden et al. 2003; Finkel et al. 2009; Cohen et al. 2004; Nemoto et al. 2004). However, it was reported recently that overexpression of SIRT1 in mice whole body does not influence lifespan (Herranz et al. 2010). In addition, recent evidence shows that overexpression of SIRT6 has a significantly increased lifespan in male mice (Kanfi et al. 2012). Nevertheless, there has been a growing recognition that sirtuins contribute to healthy lifespan by protecting against several age-associated diseases (Guarente 2011).
2.5 Mitochondria and Oxidative Damage in Yeast Replicative Lifespan
Mitochondria are semiautonomous organelles that contain their own genome (mtDNA) and have essential roles in energy production, metabolism, intracellular signaling, and apoptosis (Cheng et al. 2008; Braun and Westermann 2011; Suen et al. 2008). In addition, mitochondria are the major intracellular source of reactive oxygen species (ROS), which cause damage to cell structures and may regulate aging. However, in S. cerevisiae, the role of mitochondrial function in replicative aging is still poorly understood. In S. cerevisiae, yeasts lacking mtDNA have been associated with varying replicative lifespans depending on the strain background (Kaeberlein et al. 2005b; Kirchman et al. 1999). It has also been debated whether lifespan extension by DR requires enhanced respiration (Easlon et al. 2007; Kaeberlein et al. 2005a; Lin and Guarente 2006; Lin et al. 2000).
Similar to the lifespan phenotypes of strains lacking cytoplasmic ribosomal proteins, deletion of mitochondrial ribosomal proteins, for example, MRPL25, which encodes a component of the large subunit of the mitochondrial ribosome, leads to increased oxidative stress resistance and RLS (Heeren et al. 2009). A later study showed that strains lacking nuclear-encoded mitochondrial translation complex genes, such as SOV1, cause an Sir2-dependent RLS extension without affecting oxidative damage (Caballero et al. 2011). These studies indicate that mitochondrial translation may impinge on yeast RLS by multiple mechanisms.
Mitochondria exist in vivo as a dynamic organelle, which continually undergo fission and fusion (Westermann 2010). Recent work has shown that mitochondrial dynamics also play important roles in yeast replicative aging. A recent Mother Enrichment Program study confirmed and extended prior findings using microdissection that mitochondrial morphology changes dramatically through lifetime: mitochondria are tubular in young cells, fragmented in the early aging process, and form large aggregates in aged cells (Easlon et al. 2007; Hughes and Gottschling 2012). Mitochondria also lose membrane potential as cells age (Hughes and Gottschling 2012). The relationship between mitochondrial dynamics and aging is not fully understood. One study showed that strains lacking the mitofusin proteins Dnm1p or Fis1p, results in reduced fragmentation and extended RLS (Scheckhuber et al. 2007). Subsequently, deletion of the dynamin-related GTPases Mgm1p, results in increased fragmentation and reduced RLS and CLS (Scheckhuber et al. 2011). A recent additional study demonstrated that the double deletion of DNM1 and MGM1 exhibited a normal mitochondrial morphotype, but a decrease in mitophagy and a striking reduced RLS, indicating that a balanced mitochondrial dynamics, but not the filamentous mitochondrial morphotype might be benefit for longevity (Bernhardt et al. 2015).
The effects of mitochondrial uncouplers on yeast RLS have been studied. Partial uncoupling of oxidative phosphorylation by chemical uncoupling agent carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) results in increased levels of ROS and decreased lifespan (Stockl et al. 2007). Conversely, cells treated with dinitrophenol (DNP), which causes proton leak and uncoupling of the electron transport, results in decreased ROS and increased lifespan (Barros et al. 2004). The relationship between ROS and DR is still not clear. Recent study suggested that elevated ROS levels caused by DR result in mild stress response, which promotes lifespan extension through a hormetic mechanism (Sharma et al. 2011; Calabrese et al. 2011). Higher reactive oxygen species has also been linked to lifespan extension in worms (Yang and Hekimi 2010).
The retrograde signaling pathway mediates stress signals from the mitochondria to the nucleus. One key output of the retrograde signaling pathway is an alteration in nuclear gene expression of mitochondrial proteins, such as CIT2, in response to mitochondrial dysfunction (Epstein et al. 2001; Traven et al. 2001). It has been demonstrated that genetic and environmental manipulations that induce retrograde response pathway can increase RLS in certain strain backgrounds (Kirchman et al. 1999). Three genes have been shown to mediate the signaling pathway. RTG1 and RTG3 encode the subunits of a leucine zipper transcription factor, which activates the transcription of many retrograde response target nuclear genes for mitochondrial proteins (such as CIT2) (Rothermel et al. 1997; Jazwinski 2005a, b). RTG2 encodes part of SLIK histone acetyltransferase complex that triggers the translocation of Rtg1–Rtg3 transcription complex to the nucleus (Rothermel et al. 1995). It has been proposed that lifespan regulation by retrograde response occurs through both RTG2-independent and RTG2-dependent processes (Kirchman et al. 1999; Liu and Butow 2006). Ras-PKA and TOR pathways have been shown as upstream regulators for the retrograde signaling (Tate and Cooper 2003; Kirchman et al. 1999; Borghouts et al. 2004; Komeili et al. 2000). However, another study indicated that activation of retrograde response by mitochondria leads to increased ERC accumulation (Borghouts et al. 2004). Overall, the mechanisms by which the retrograde response regulates lifespan in yeast remain unclear; however, it is of interest because the mitochondria-to-nucleus signaling pathway may have an evolutionarily conserved role in the aging process in higher organisms (Kujoth et al. 2007; Sedensky and Morgan 2006).
Recent studies on mitochondria have elucidated a link between mitochondrial dysfunction and vacuolar acidification (Hughes and Gottschling 2012). Vacuolar acidity decreases during the aging process, which leads to mitochondrial dysfunction through reducing pH-dependent amino acid storage in the vacuolar lumen (Hughes and Gottschling 2012). Preventing the decline of vacuolar acidity leads to a stabilization of mitochondrial function and increased lifespan (Hughes and Gottschling 2012). Along similar lines, it has been illustrated that increased vacuolar fusion, by overexpression of OSH6, leads to increased lifespan (Gebre et al. 2012). With multiple lines of evidence pointing to a linked function of the vacuole and mitochondria related to aging in yeast (Hughes and Gottschling 2012), it will be important to determine which of these events are conserved in mammalian aging, as well as adult stem cell function.
2.6 The Ubiquitin/Proteasome System in Yeast Replicative Lifespan
It has also been reported that the ubiquitin/proteasome system (UPS) may play a crucial role in RLS (Dange et al. 2011; Kruegel et al. 2011). Importantly, proteasome activity acts as a central regulator of oxidative stress and aging in different organisms, providing strong evidence for a conserved potential effect on longevity from yeast to humans (Vernace et al. 2007; Carrard et al. 2002; Ghazi et al. 2007). Long-lived organisms such as the naked mole rat display higher levels of proteasome activity when compared to similar shorter-lived species (Perez et al. 2009). In addition, a recent study showed that germline ablation in C. elegans results in increased somatic UPS activity and lifespan extension (Vilchez et al. 2012).
The function of UPS decreases during aging (Shringarpure and Davies 2002; Baraibar and Friguet 2012). Importantly, increased UPS activity results in a significantly increased RLS and resistance to proteotoxic stress (Kruegel et al. 2011). It has been proposed that increased proteasome activity is beneficial for longevity and is due to improved elimination of damaged proteins which are asymmetrically segregated to the mother cell during cell division (Zhou et al. 2011; Kruegel et al. 2011). Recently, a study tried to answer why UPS activity declines as cells age. If the level of damaged proteins reaches the proteasome capacity, then protein aggregates will occur. However, as cells age, aggregated proteins appear to accumulate beyond a threshold whereby they interfere directly with proteasome function creating a negative feedback loop (Andersson et al. 2013). A new study demonstrated that proteasomes regulates the AMPK pathway by targeting Mig1, a transcription factor involved in glucose repression (Yao et al. 2015). Increased proteasome activity caused reduced Mig1 level and incorrect localization of Mig1 (Yao et al. 2015). These findings might impact not only mammalian aging, but a wide range of protein aggregation-based diseases.
A recent study reported that yeast metacaspase (Mca1) play a critical role in aggregate management and RLS (Hill et al. 2014). MCA1 showed to help degrade misfolded proteins that left into the daughter cells. MCA1 deletion didn’t affect lifespan in wild type strains, but decreased RLS in strains lacking Ydj1, the Hsp70 co-chaperone. Subsequently, overexpressing MCA1 increased RLS in YDJ1 deletion background. These findings not only challenges the idea that caspases execute cellular suicide in an altruistic mechanism, but also might shed lights on protein degradation and rejuvenation mechanisms in stem cells (Kampinga, 2014) (Hill and Nystrom, 2015).
2.7 The Methodology of Chronological Lifespan and New Variants
Chronological lifespan (CLS) is the length of time that a population of nondividing yeast cells can maintain viability or replicative potential (Longo et al. 1996). The chronological lifespan assay was developed as an alternative to the highly tedious replicative lifespan assays. Rather than measuring lifespan in a mitotic state, the chronological lifespan assay does so in a postmitotic or quiescent state. This has been argued to be a better representation of nondividing cells in higher organisms, but also has drawbacks that are addressed later in this section. Unlike the RLS studies, the CLS assay is more conducive to biochemical studies since survival is assessed in a population of cells.
Yeast cells are facultative anaerobes, generating energy mostly through fermentation in rich media with high glucose levels, but switching to respiration when nutrients become scarce. After nutrient depletion, cells enter a nonproliferative stress-resistant state where they can retain viability for a period from days to weeks depending on media conditions. While diploid cells can sporulate under certain conditions, the CLS assay is performed typically in haploid cells or in diploids in a non-sporulation environment. The most common method for CLS is to culture cells in liquid synthetic defined media for a period of 2–3 days where they progress through a postdiauxic, respiratory state and reach maximum cell density before stopping proliferation and ultimately stationary phase. Then, the percentage of cells that retain viability are monitored by periodically plating serial dilutions onto rich media and monitoring colony formation (Kaeberlein 2006; Fabrizio and Longo 2007). Readers can refer to these citations for detailed written and video protocols for a high-throughput method using a Bioscreen machine to obtain outgrowth curves from aged cells (Murakami et al. 2008; Murakami and Kaeberlein 2009). In addition, tools such as Yeast Outgrowth Data Analysis (YODA) have been developed in order to assist researchers (Olsen et al. 2010).
There are several variations of the chronological lifespan protocol, with each having its own strengths and weaknesses. All the methods are variations on the concept of placing cells in a postmitotic state and measuring how long the cells are able to maintain viability (Longo et al. 1996). This method attempts to mimic the conditions that yeast may commonly encounter in their environment, but slight variations in the preparation of the cultures such as the type of caps used on the culture tubes and the media/flask volume ratios can drastically alter the aeration of the culture, directly altering viability and influencing the pathways that modulate lifespan (Hu et al. 2014; Longo et al. 2012).
Several other complementary CLS methods have been developed. One involves measuring viability after starting cultures in a calorie-restricted state and another starting cells in normal conditions, but transferring them from spent media to water after nutrients are depleted. Both methods drastically extend chronologic lifespan. Depending on growth conditions, buildup of acetic acid during the growth phase as a result of fermentation can acidify media and accelerate cell death in the nonproliferative phase (Burtner et al. 2009). This observation has also been demonstrated to be true in several strain backgrounds, as well as wild vineyard strains (Murakami et al. 2011). This is one reason why starting cells in reduced glucose or transferring them to water after proliferation ceases extends lifespan. However, by altering growth conditions, the impact of acetic acid can be reduced (Hu et al. 2014; Longo et al. 2012). One additional way to reduce effects of acetic acid is to grow yeast in buffered media, so that the pH does not drop with acetic acid accumulation and secretion (Burtner et al. 2009).
Another method has been developed where cells in a tryptophan auxotrophic background are plated onto solid media lacking tryptophan. At every time point, tryptophan is added to the plate, and the number of colonies that form is counted (Wei et al. 2009). This method, however, is less conducive to high-throughput. In addition, measuring lifespan by this method slightly reduces chronological lifespan compared to the liquid media approach. The differences can likely be accounted for by the differential response of yeast cells depending on the nutrients for which they are limited. Cells starving for carbohydrates respond differently than tryptophan, for instance. An analysis of the response of yeast under a variety of conditions indicates that the cellular response depends on the first nutrient that becomes depleted (Gresham et al. 2011). Cells encountering starvation in a condition for which they are adapted to respond (e.g., a nutrient for which they would normally become restricted in the wild) have extended survival and evoke a common stress response that protects against a wide range of stressing agents. However, starvation in an unnatural condition leads to an ineffective response and rapid loss of viability.
2.8 Genes and Pathways Modulating Chronological Lifespan
By using a combination of several CLS methods, it has been possible to identify genetic and environmental manipulations that regulate chronological lifespan. Dietary restriction, which for CLS is defined as growth under reduced glucose, typically at a concentration of 0.05 % of the culture medium, has been reliably demonstrated to extend chronological aging significantly (Smith et al. 2007). Similarly, transfer of postmitotic cells to water also extends chronological lifespan. DR has been found to increase respiration (Lin et al. 2002), as a part of a global restructuring of the metabolic state of the cells. Consistent with this hypothesis, cells grown under caloric restriction result in a more basic medium, whereas cells grown under ordinary culture conditions (2 % w/v glucose) actually acidify their medium modestly. The difference in pH in the media likely is a result of byproducts of the metabolic restructuring, as Burtner et al. demonstrated that acetic acid is produced during growth and expelled into the extracellular environment (Burtner et al. 2009). Similarly, Burtner et al. demonstrated that acetic acid is a likely candidate because in cultures grown under calorie restriction or under 3 % glycerol, which forces the cells to respire, because the culture medium was not acidified and acetic acid did not accumulate under these conditions. Interestingly, a similar phenomenon has been reported for growth of mammalian cells in culture. Cells grown in culture acidify their medium by secretion of lactic acid, which reduces their long-term viability. Interestingly, rapamycin extends survival by reducing lactic acid production (Leontieva and Blagosklonny 2011).
Consistent with the findings that altered metabolism extends chronological lifespan, the two main pathways that have been well characterized for extending chronological lifespan are the nutrient sensing PKA and TOR/SCH9 pathways. Genetic perturbations in both have been demonstrated to reliably extend chronological lifespan in a variety of chronological methods, and these effects have been pinpointed to be a result of Rim15 and its downstream effectors Msn2/4 (Burtner et al. 2009). In accordance, reduced Tor signaling increases mitochondrial translation, oxidative phosphorylation (OXPHOS), and mitochondrial respiration, as well as mitochondrial reactive oxygen species (Bonawitz et al. 2007; Pan and Shadel 2009). These results taken together suggest that chronological aging is a result of an adaptive mitochondrial response, especially since stimulation of ROS during the growth phase is sufficient to extend chronological lifespan. In addition, pre-growth on glycerol and HAP4 overexpression, which activates respiration, both extends CLS (Piper et al. 2006).
One of the most studied members of the Tor pathway is SCH9. Consistent with its direct role in the Tor pathway, several studies have demonstrated that deletion of SCH9 extends chronological lifespan. Also consistent with the metabolic perturbations observed of cells under calorie restriction, sch9Δ demonstrates metabolic alterations, specifically enhanced glycerol biosynthesis. Glycerol biosynthetic genes are upregulated in sch9Δ, and these genes are required for the chronological lifespan extension in sch9Δ (Wei et al. 2009). They also proposed that glycerol is an important stress protectant during chronological aging. Cells lacking SCH9 are also protected from loss of viability due to extracellular acetic acid (Burtner et al. 2009). tor1D cells also exhibit enhanced glycerol biosynthesis (Wei et al. 2009).
Another mechanism that has been directly linked to chronological lifespan is the role that membrane potential of the mitochondria plays in chronological aging. It is documented that as cells age by replication, their mitochondria display signs of altered morphology and of decreased membrane potential (Easlon et al. 2007; Hughes and Gottschling 2012). Respiration is also required for normal chronological lifespan; however, up to a 60 % reduction in respiration does not affect lifespan (Pan et al. 2011; Ocampo et al. 2012). Overexpression of superoxide dismutases curtails the extended chronological lifespan of Tor mutants (Pan et al. 2011), consistent with the finding that reactive oxygen species can promote enhanced CLS (Piper et al. 2006). These observations, along with the fact that addition of trehalose to the media is sufficient to restore chronological aging in respiratory-deficient mutants, suggest that there is an elevated stress response that is dependent on reactive oxygen species. Although yet to be fully elucidated, this mitochondrial reactive oxygen species signaling could prove a fruitful pathway to study given the multitude of diseases associated with aberrant mitochondrial function.
2.9 RLS and CLS: From Yeast to Mammalian Aging
Since the earliest chronological lifespan studies, it has been known that several interventions that affect chronological aging overlap with those that also affect replicative aging, such as DR, TOR/Sch9 pathway, and PKA pathway (Lin et al. 2000; Fabrizio et al. 2001, 2004). Mitochondria function and oxidative damage are also associated with both replicative and chronological aging (Kaeberlein 2010). However, some mutations have been reported to have conflicting effects in RLS and CLS assays (Harris et al. 2001, 2003; Fabrizio et al. 2004). rDNA instability is relevant to RLS, but not CLS since rDNA instability by sir2Δ results does not affect CLS (Smith et al. 2007; Wu et al. 2011; Kennedy et al. 2005).
Recently, a direct link between the CLS and RLS assays has been elucidated. Delaney et al. studied the effects chronological aging on the subsequent RLS of mother cells (Delaney et al. 2013). They demonstrated, consistent with an earlier report (Ashrafi et al. 1999), that cells aged chronologically display a reduced replicative potential and that this is exacerbated under conditions that give rise to acetic acid toxicity (Delaney et al. 2013).
Chronological aging and replicative aging have both shed light on similar pathways and have been insightful on their mechanisms. The first quantitative analysis study to address if the longevity genes are conserved between S. cerevisiae and C. elegans was demonstrated by Smith et al. (Smith et al. 2008). They found that the yeast orthologs of worm long-lived genes are more likely to impact yeast RLS, indicating that genes modulating aging have been conserved between divergent eukaryotic species (Smith et al. 2008). On an evolutionary timeline, the unicellular yeast S. cerevisiae and multicellular worm C. elegans are separated by 1.5 billion years, which is greater than worm and mammals (1 billion years) (Wang et al. 1999). This indicates that the conserved genes between yeast and worm are likely to have similar overlap in mammals. In contrast to RLS, and in spite of the fact that many of the major pathways affecting RLS, CLS, and worm aging are conserved, there has been no successful quantitative demonstration of conservation in aging pathways between yeast CLS and worm aging or even between CLS and RLS. The efforts to date have centered on CLS conditions where acidification is a prime determinant of aging (Burtner et al. 2011), and it may be the case that other methods of CLS may lead to greater correspondence with aging determinants in the RLS assay and multicellular eukaryotes.
Both the replicative aging and chronological aging assays are meant to establish aging mechanisms in relatively simple organisms that can be tested in metazoans. Chronological aging is proposed as a model for postmitotic cells such as neurons, while replicative aging can be a model for dividing populations of cells, such as adult stem cells. Like budding yeast, it has been reported that damaged proteins in stem cells are asymmetrically distributed during cell division (Fuentealba et al. 2008; Rujano et al. 2006), although it remains unclear whether the mechanisms by which this segregation occurs are conserved in mammals. We predict that the budding yeast replicative aging studies will provide further insights into key regulators for stem cell development and senescence (Thorpe et al. 2008).
In addition, yeast is increasingly used to model aging-related disease, such as neurodegenerative diseases and cancer (we refer readers with specific interest in disease models to a dedicated review (Tenreiro et al. 2013; Pereira et al. 2012)). For example, Werner syndrome is an autosomal recessive disorder characterized by the appearance of premature aging and cancer predisposition (Epstein et al. 1966). Werner syndrome is caused by mutation of the WRN gene, which functions in telomere maintenance, DNA replication, and DNA repair (Yu et al. 1996). It has been shown that deletion of WRN ortholog in yeast, SGS1, results in decreased lifespan and elevated ERC accumulation (McVey et al. 2001). Many other chronic diseases have also been modeled.
The yeast model not only has provided a valuable model to discover compounds that impact aging. Many compounds have now been identified that extend yeast lifespan (Table 2.1). Examples include rapamycin and resveratrol, which were first identified and characterized as antiaging compounds in yeast and have been studied for a number of aging-related diseases in humans (Lamming et al. 2013; Kennedy and Pennypacker 2014). Novel targets such as serine palmitoyltransferase (the first enzyme in the de novo sphingolipid biosynthesis pathway) have also been identified that play important roles in aging, providing further promising pharmacological targets for antiaging interventions (Huang et al. 2014). Moreover, a systems biology approach has been applied to yeast to identify perturbations that can potentially transform an “aged” metabolic state to the desired “young” metabolic state, as well as predict new antiaging drug targets (Yizhak et al. 2013).
Table 2.1
Compounds increase lifespan in S. cerevisiae
Compound | Model | Type of compound | Potential mechanism | References |
|---|---|---|---|---|
Rapamycin | RLS, CLS | Natural product, immunosuppressant | TOR | |
Resveratrol | RLS | Polyphenol, natural product in red wine | Stimulate Sir2, increase DNA stability | Howitz et al. (2003) |
Ibuprofen | RLS | nonsteroidal anti-inflammatory drug | Tryptophan import | He et al. (2014) |